Gaussian Assumption for Normality
The Gaussian (normal) distribution is a key assumption in many statistical methods. It is mathematically represented by the probability density function (PDF):
where:
\(\mu\) is the mean
\(\sigma^2\) is the variance
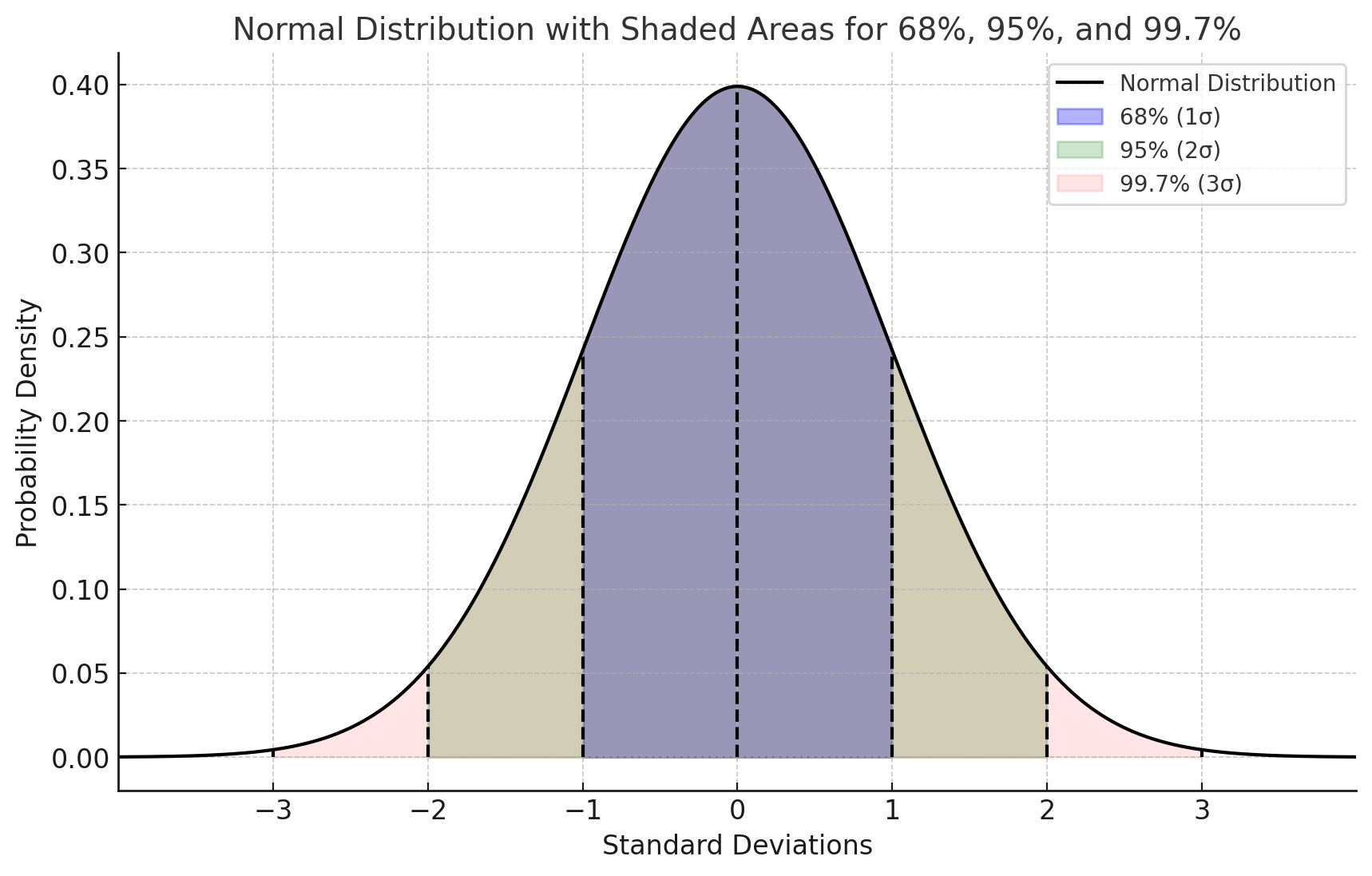
In a normally distributed dataset:
68% of data falls within \(\mu \pm \sigma\)
95% within \(\mu \pm 2\sigma\)
99.7% within \(\mu \pm 3\sigma\)
Histograms and Kernel Density Estimation (KDE)
Histograms:
Visualize data distribution by binning values and counting frequencies.
If data is Gaussian, the histogram approximates a bell curve.
KDE:
A non-parametric way to estimate the PDF by smoothing individual data points with a kernel function.
The KDE for a dataset \(X = \{x_1, x_2, \ldots, x_n\}\) is given by:
where:
\(K\) is the kernel function (often Gaussian)
\(h\) is the bandwidth (smoothing parameter)
Histograms offer a discrete, binned view of the data.
KDE provides a smooth, continuous estimate of the underlying distribution.
Together, they effectively illustrate how well the data aligns with the Gaussian assumption, highlighting any deviations from normality.
Pearson Correlation Coefficient
The Pearson correlation coefficient, often denoted as \(r\), is a measure of the linear relationship between two variables. It quantifies the degree to which a change in one variable is associated with a change in another variable. The Pearson correlation ranges from \(-1\) to \(1\), where:
\(r = 1\) indicates a perfect positive linear relationship.
\(r = -1\) indicates a perfect negative linear relationship.
\(r = 0\) indicates no linear relationship.
The Pearson correlation coefficient between two variables \(X\) and \(Y\) is defined as:
where:
\(\text{Cov}(X, Y)\) is the covariance of \(X\) and \(Y\).
\(\sigma_X\) is the standard deviation of \(X\).
\(\sigma_Y\) is the standard deviation of \(Y\).
Covariance measures how much two variables change together. It is defined as:
where:
\(n\) is the number of data points.
\(X_i\) and \(Y_i\) are the individual data points.
\(\mu_X\) and \(\mu_Y\) are the means of \(X\) and \(Y\).
The standard deviation measures the dispersion or spread of a set of values. For a variable \(X\), the standard deviation \(\sigma_X\) is:
Substituting the covariance and standard deviation into the Pearson correlation formula:
This formula normalizes the covariance by the product of the standard deviations of the two variables, resulting in a dimensionless coefficient that indicates the strength and direction of the linear relationship between \(X\) and \(Y\).
\(r > 0\): Positive correlation. As \(X\) increases, \(Y\) tends to increase.
\(r < 0\): Negative correlation. As \(X\) increases, \(Y\) tends to decrease.
\(r = 0\): No linear correlation. There is no consistent linear relationship between \(X\) and \(Y\).
The closer the value of \(r\) is to \(\pm 1\), the stronger the linear relationship between the two variables.
Box-Cox Transformation
The Box-Cox transformation is a powerful technique for stabilizing variance and making data more closely follow a normal distribution. Developed by statisticians George Box and David Cox in 1964, the transformation is particularly useful in linear regression models where assumptions of normality and homoscedasticity are necessary. This document provides an accessible overview of the theoretical concepts underlying the Box-Cox transformation.
Many statistical methods assume that data is normally distributed and that the variance remains constant across observations (homoscedasticity). However, real-world data often violates these assumptions, especially when dealing with positive-only, skewed distributions (e.g., income, expenditure, biological measurements). The Box-Cox transformation is a family of power transformations designed to address these issues by “normalizing” the data and stabilizing variance.
Mathematical Definition
The Box-Cox transformation is defined as follows:
Here:
\(y(\lambda)\) is the transformed variable,
\(y\) is the original variable (positive and continuous),
\(\lambda\) is the transformation parameter.
When \(\lambda = 0\), the transformation becomes a natural logarithm, effectively a special case of the Box-Cox transformation.
Interpretation of the Lambda Parameter
The value of \(\lambda\) determines the shape of the transformation:
\(\lambda = 1\): The transformation does nothing; the data remains unchanged.
\(\lambda = 0.5\): A square-root transformation.
\(\lambda = 0\): A logarithmic transformation.
\(\lambda < 0\): An inverse transformation, which is often helpful when working with highly skewed data.
Selecting the optimal value of \(\lambda\) to achieve approximate normality or homoscedasticity is typically done using maximum likelihood estimation (MLE), where the goal is to find the value of \(\lambda\) that maximizes the likelihood of observing the transformed data under a normal distribution.
Properties and Benefits
The Box-Cox transformation has two key properties:
Variance Stabilization: By choosing an appropriate \(\lambda\), the variance of \(y(\lambda)\) can be made more constant across levels of \(y\). This is particularly useful in regression analysis, as homoscedasticity is often a critical assumption.
Normalization: The transformation makes the distribution of \(y(\lambda)\) closer to normality. This allows statistical techniques that assume normality to be more applicable to real-world, skewed data.
Likelihood Function
The likelihood function for the Box-Cox transformation is derived from the assumption that the transformed data follows a normal distribution. For a dataset with observations \(y_i\), the likelihood function is given by:
where:
\(n\) is the number of observations,
\(s^2\) is the sample variance of the transformed data.
Maximizing this likelihood function provides the MLE for \(\lambda\), which can be estimated using iterative methods.
Practical Considerations
In practice, implementing the Box-Cox transformation requires a few considerations:
Positive-Only Data: The transformation is only defined for positive values. For datasets with zero or negative values, a constant can be added to make all observations positive before applying the transformation.
Interpretability: The transformed data may lose interpretability in its original scale. For some applications, this trade-off is justified to meet model assumptions.
Inverse Transformation: If interpretability is a concern, the inverse of the Box-Cox transformation can be applied to transform results back to the original scale.
Applications in Modeling
In regression modeling, the Box-Cox transformation can improve both the accuracy and validity of predictions. For example, in Ordinary Least Squares (OLS) regression, the transformation reduces heteroscedasticity and normalizes residuals, leading to more reliable parameter estimates. Similarly, in time series analysis, the Box-Cox transformation can stabilize variance, making models such as ARIMA more effective.
The Box-Cox transformation is a flexible and powerful technique for addressing non-normality and heteroscedasticity in data. By choosing an appropriate \(\lambda\), practitioners can transform data to better meet the assumptions of various statistical methods, enhancing the reliability of their models and inferences.
Confidence Intervals for Lambda
In practice, it is often helpful to assess the stability of the estimated transformation parameter \(\lambda\) by constructing a confidence interval (CI). The CI provides a range of values within which the true value of \(\lambda\) is likely to fall, offering insights into the sensitivity of the transformation.
To construct a confidence interval for \(\lambda\), the following approach can be used:
1. Alpha Level: Select an alpha level, commonly 0.05, for a 95% confidence interval, or adjust as needed. The alpha level represents the probability of observing a value outside this interval if the estimate were repeated multiple times.
2. Profile Likelihood Method: One approach is to use the profile likelihood method, where a range of \(\lambda\) values are tested, and those with likelihoods close to the maximum likelihood estimate (MLE) are retained within the interval. The confidence interval is defined as the set of \(\lambda\) values for which the likelihood ratio statistic:
\[\text{LR}(\lambda) = 2 \left( L(\hat{\lambda}) - L(\lambda) \right)\]is less than the chi-square value at the chosen confidence level (e.g., 3.84 for a 95% CI with one degree of freedom).
Interpretation: A narrow CI around \(\lambda\) suggests that the transformation is relatively stable, while a wide interval might indicate sensitivity, signaling that the data may benefit from an alternative transformation or modeling approach.
These confidence intervals provide a more robust understanding of the transformation’s impact, as well as the degree of transformation needed to meet model assumptions.
The Yeo-Johnson Transformation
For a feature \(y\), the Yeo-Johnson transformation \(Y\) is defined as:
Breakdown of the Conditions
- For Positive Values of \(y\) (\(y \geq 0\)):
When \(\lambda \neq 0\): The transformation behaves similarly to the Box-Cox transformation with \((y + 1)^{\lambda}\).
When \(\lambda = 0\): The transformation simplifies to the natural log, \(\ln(y + 1)\).
- For Negative Values of \(y\) (\(y < 0\)):
When \(\lambda \neq 2\): A reflected transformation is applied, \(-(−y + 1)^{2 - \lambda}\), to manage negative values smoothly.
When \(\lambda = 2\): The transformation simplifies to \(- \ln(-y + 1)\), making it suitable for negative inputs while preserving continuity.
Why It Works
The Yeo-Johnson transformation adjusts data to make it more normally distributed. By allowing transformations for both positive and negative values, it offers flexibility across various distributions. The parameter \(\lambda\) is typically optimized to best approximate normality.
When to Use It
Yeo-Johnson is particularly useful for datasets containing zero or negative values. It’s often effective for linear models that assume normally distributed data, making it a robust alternative when Box-Cox cannot be applied.
Median and IQR Scaling
RobustScaler in scikit-learn is a scaling method that reduces the impact
of outliers in your data by using the median and interquartile range (IQR)
instead of the mean and standard deviation, which are more sensitive to extreme values.
Here’s a mathematical breakdown of how it works:
Centering Data Using the Median
The formula for scaling each feature \(x\) in the dataset using RobustScaler is:
where:
\(\text{Median}(x)\) is the median of the feature \(x\).
\(\text{IQR}(x) = Q_3 - Q_1\), the interquartile range, is the difference between the 75th percentile (\(Q_3\)) and the 25th percentile (\(Q_1\)) of the feature. This range represents the spread of the middle 50% of values, which is less sensitive to extreme values than the total range.
Explanation of Each Component
Median (\(\text{Median}(x)\)): This is the 50th percentile, or the central value of the feature. It acts as the “center” of the data, but unlike the mean, it is robust to outliers.
Interquartile Range (IQR): By dividing by the IQR, the
RobustScalerstandardizes the spread of the data based on the range of the middle 50% of values, making it less influenced by extreme values. Essentially, the values are scaled to fall within a range close to -1 to 1 for the majority of samples.
Example Calculation
Suppose you have a feature \(x = [1, 2, 3, 4, 5, 100]\). Here’s how the scaling would work:
Calculate the Median:
\[\text{Median}(x) = 3.5\]Calculate the Interquartile Range (IQR):
First, find \(Q_1\) (25th percentile) and \(Q_3\) (75th percentile): - \(Q_1 = 2\), \(Q_3 = 5\)
Then, \(\text{IQR}(x) = Q_3 - Q_1 = 5 - 2 = 3\)
Apply the Scaling Formula:
For each \(x\) value, subtract the median and divide by the IQR:
\[x_{\text{scaled}} = \frac{x - 3.5}{3}\]
This results in values that are centered around 0 and scaled according to the interquartile range, rather than the full range or mean and standard deviation. For our example, the outlier (100) will be downscaled effectively, reducing its influence on the data’s range and making the scaling robust to such outliers.
The RobustScaler is particularly useful when dealing with data with significant
outliers, as it centers the data around the median and scales according to the
IQR, allowing for better handling of extreme values than traditional
standardization methods.
Logit Transformation
The logit transformation is used to map values from the range \((0, 1)\) to the entire real number line \((-\infty, +\infty)\). It is defined mathematically as:
where \(p\) is a value in the range \((0, 1)\). In other words, for each value \(p\), the transformation is calculated by taking the natural logarithm of the odds \(p / (1 - p)\).
Purpose and Assumptions
The logit function is particularly useful in scenarios where data is constrained between 0 and 1, such as probabilities or proportions. However, to apply this transformation, all values must strictly lie within the open interval \((0, 1)\). Values equal to 0 or 1 result in undefined values \((-\infty, +\infty\) respectively) since the logarithm of zero is undefined.
In the code implementation, a ValueError is raised if any values in the target feature fall outside the
interval \((0, 1)\). If your data does not meet this condition, consider applying a Min-Max scaling first to transform
the data to the appropriate range.
Example
If \(p = 0.5\), then: