Creating Effective Visualizations
This section focuses on practical strategies and techniques for designing clear and impactful visualizations using the diverse plotting tools provided in the EDA Toolkit.
Heuristics for Visualizations
When creating visualizations, there are several key heuristics to keep in mind:
Clarity: The visualization should clearly convey the intended information without ambiguity.
Simplicity: Avoid overcomplicating visualizations with unnecessary elements; focus on the data and insights.
Consistency: Ensure consistent use of colors, shapes, and scales across visualizations to facilitate comparisons.
Methodologies
The EDA Toolkit supports the following methodologies for creating effective visualizations:
KDE and Histograms Plots: Useful for showing the distribution of a single variable. When combined, these can provide a clearer picture of data density and distribution.
Feature Scaling and Outliers: Identifying outliers is critical for understanding the data’s distribution and potential anomalies. The EDA Toolkit offers various methods for outlier detection, including enhanced visualizations using box plots and scatter plots.
Stacked Crosstab Plots: These are used to display multiple data series on the same chart, comparing cumulative quantities across categories. In addition to the visual stacked bar plots, the corresponding crosstab table is printed alongside the visualization, providing detailed numerical insight into how the data is distributed across different categories. This combination allows for both a visual and tabular representation of categorical data, enhancing interpretability.
Box and Violin Plots: Useful for visualizing the distribution of data points, identifying outliers, and understanding the spread of the data. Box plots are particularly effective when visualizing multiple categories side by side, enabling comparisons across groups. Violin plots provide additional insights by showing the distribution’s density, giving a fuller picture of the data’s distribution shape.
Scatter Plots and Best Fit Lines: Effective for visualizing relationships between two continuous variables. Scatter plots can also be enhanced with regression lines or trend lines to identify relationships more clearly.
Correlation Matrices: Helpful for visualizing the strength of relationships between multiple variables. Correlation heatmaps use color gradients to indicate the degree of correlation, with options for annotating the values directly on the heatmap.
Partial Dependence Plots: Useful for visualizing the relationship between a target variable and one or more features after accounting for the average effect of the other features. These plots are often used in model interpretability to understand how specific variables influence predictions.
KDE and Histogram Distribution Plots
KDE Distribution Function
Generate KDE or histogram distribution plots for specified columns in a DataFrame.
The kde_distributions function is a versatile tool designed for generating
Kernel Density Estimate (KDE) plots, histograms, or a combination of both for
specified columns within a DataFrame. This function is particularly useful for
visualizing the distribution of numerical data across various categories or groups.
It leverages the powerful seaborn library [2] for plotting, which is built on top of
matplotlib [3] and provides a high-level interface for drawing attractive and informative
statistical graphics.
Key Features and Parameters
Flexible Plotting: The function supports creating histograms, KDE plots, or a combination of both for specified columns, allowing users to visualize data distributions effectively.
Leverages Seaborn Library: The function is built on the seaborn library, which provides high-level, attractive visualizations, making it easy to create complex plots with minimal code.
Customization: Users have control over plot aesthetics, such as colors, fill options, grid sizes, axis labels, tick marks, and more, allowing them to tailor the visualizations to their needs.
Scientific Notation Control: The function allows disabling scientific notation on the axes, providing better readability for certain types of data.
Log Scaling: The function includes an option to apply logarithmic scaling to specific variables, which is useful when dealing with data that spans several orders of magnitude.
Output Options: The function supports saving plots as PNG or SVG files, with customizable filenames and output directories, making it easy to integrate the plots into reports or presentations.
- kde_distributions(df, vars_of_interest=None, figsize=(5, 5), grid_figsize=None, hist_color='#0000FF', kde_color='#FF0000', mean_color='#000000', median_color='#000000', hist_edgecolor='#000000', hue=None, fill=True, fill_alpha=1, n_rows=None, n_cols=None, w_pad=1.0, h_pad=1.0, image_path_png=None, image_path_svg=None, image_filename=None, bbox_inches=None, single_var_image_filename=None, y_axis_label='Density', plot_type='both', log_scale_vars=None, bins='auto', binwidth=None, label_fontsize=10, tick_fontsize=10, text_wrap=50, disable_sci_notation=False, stat='density', xlim=None, ylim=None, plot_mean=False, plot_median=False, std_dev_levels=None, std_color='#808080', label_names=None, show_legend=True, **kwargs)
- Parameters:
df (pandas.DataFrame) – The DataFrame containing the data to plot.
vars_of_interest (list of str, optional) – List of column names for which to generate distribution plots. If ‘all’, plots will be generated for all numeric columns.
figsize (tuple of int, optional) – Size of each individual plot, default is
(5, 5). Used when only one plot is being generated or when saving individual plots.grid_figsize (tuple of int, optional) – Size of the overall grid of plots when multiple plots are generated in a grid. Ignored when only one plot is being generated or when saving individual plots. If not specified, it is calculated based on
figsize,n_rows, andn_cols.hist_color (str, optional) – Color of the histogram bars, default is
'#0000FF'.kde_color (str, optional) – Color of the KDE plot, default is
'#FF0000'.mean_color (str, optional) – Color of the mean line if
plot_meanis True, default is'#000000'.median_color (str, optional) – Color of the median line if
plot_medianis True, default is'#000000'.hist_edgecolor (str, optional) – Color of the histogram bar edges, default is
'#000000'.hue (str, optional) – Column name to group data by, adding different colors for each group.
fill (bool, optional) – Whether to fill the histogram bars with color, default is
True.fill_alpha (float, optional) – Alpha transparency for the fill color of the histogram bars, where
0is fully transparent and1is fully opaque. Default is1.n_rows (int, optional) – Number of rows in the subplot grid. If not provided, it will be calculated automatically.
n_cols (int, optional) – Number of columns in the subplot grid. If not provided, it will be calculated automatically.
w_pad (float, optional) – Width padding between subplots, default is
1.0.h_pad (float, optional) – Height padding between subplots, default is
1.0.image_path_png (str, optional) – Directory path to save the PNG image of the overall distribution plots.
image_path_svg (str, optional) – Directory path to save the SVG image of the overall distribution plots.
image_filename (str, optional) – Filename to use when saving the overall distribution plots.
bbox_inches (str, optional) – Bounding box to use when saving the figure. For example,
'tight'.single_var_image_filename (str, optional) – Filename to use when saving the separate distribution plots. The variable name will be appended to this filename. This parameter uses
figsizefor determining the plot size, ignoringgrid_figsize.y_axis_label (str, optional) – The label to display on the
y-axis, default is'Density'.plot_type (str, optional) – The type of plot to generate, options are
'hist','kde', or'both'. Default is'both'.log_scale_vars (str or list of str, optional) – Variable name(s) to apply log scaling. Can be a single string or a list of strings.
bins (int or sequence, optional) – Specification of histogram bins, default is
'auto'.binwidth (float, optional) – Width of each bin, overrides bins but can be used with binrange.
label_fontsize (int, optional) – Font size for axis labels, including xlabel, ylabel, and tick marks, default is
10.tick_fontsize (int, optional) – Font size for tick labels on the axes, default is
10.text_wrap (int, optional) – Maximum width of the title text before wrapping, default is
50.disable_sci_notation (bool, optional) – Toggle to disable scientific notation on axes, default is
False.stat (str, optional) – Aggregate statistic to compute in each bin (e.g.,
'count','frequency','probability','percent','density'), default is'density'.xlim (tuple or list, optional) – Limits for the
x-axisas a tuple or list of (min,max).ylim (tuple or list, optional) – Limits for the
y-axisas a tuple or list of (min,max).plot_mean (bool, optional) – Whether to plot the mean as a vertical line, default is
False.plot_median (bool, optional) – Whether to plot the median as a vertical line, default is
False.std_dev_levels (list of int, optional) – Levels of standard deviation to plot around the mean.
std_color (str or list of str, optional) – Color(s) for the standard deviation lines, default is
'#808080'.label_names (dict, optional) – Custom labels for the variables of interest. Keys should be column names, and values should be the corresponding labels to display.
show_legend (bool, optional) – Whether to show the legend on the plots, default is
True.kwargs (additional keyword arguments) – Additional keyword arguments passed to the Seaborn plotting function.
- Raises:
If
plot_typeis not one of'hist','kde', or'both'.If
statis not one of'count','density','frequency','probability','proportion','percent'.If
log_scale_varscontains variables that are not present in the DataFrame.If
fillis set toFalseandhist_edgecoloris not the default.If
grid_figsizeis provided when only one plot is being created.
If both
binsandbinwidthare specified, which may affect performance.
- Returns:
None
Notes
If you do not set n_rows or n_cols to any values, the function will
automatically calculate and create a grid based on the number of variables being
plotted, ensuring an optimal arrangement of the plots.
To save images, the paths for image_path_png or image_path_svg must be specified.
The trigger for saving plots is the presence of image_filename as a string.
KDE and Histograms Example
In the below example, the kde_distributions function is used to generate
histograms for several variables of interest: "age", "education-num", and
"hours-per-week". These variables represent different demographic and
financial attributes from the dataset. The plot_type="both" parameter ensures that a
Kernel Density Estimate (KDE) plot is overlaid on the histograms, providing a
smoothed representation of the data’s probability density.
The visualizations are arranged in a single row of four columns, as specified
by n_rows=1 and n_cols=3, respectively. The overall size of the grid
figure is set to 14 inches wide and 4 inches tall (grid_figsize=(14, 4)),
while each individual plot is configured to be 4 inches by 4 inches
(single_figsize=(4, 4)). The fill=True parameter fills the histogram
bars with color, and the spacing between the subplots is managed using
w_pad=1 and h_pad=1, which add 1 inch of padding both horizontally and
vertically.
To handle longer titles, the text_wrap=50 parameter ensures that the title
text wraps to a new line after 50 characters. The bbox_inches="tight" setting
is used when saving the figure, ensuring that it is cropped to remove any excess
whitespace around the edges. The variables specified in vars_of_interest are
passed directly to the function for visualization.
Each plot is saved individually with filenames that are prefixed by
"kde_density_single_distribution", followed by the variable name. The `y-axis`
for all plots is labeled as “Density” (y_axis_label="Density"), reflecting that
the height of the bars or KDE line represents the data’s density. The histograms
are divided into 10 bins (bins=10), offering a clear view of the distribution
of each variable.
Additionally, the font sizes for the axis labels and tick labels
are set to 16 points (label_fontsize=16) and 14 points (tick_fontsize=14),
respectively, ensuring that all text within the plots is legible and well-formatted.
from eda_toolkit import kde_distributions
vars_of_interest = [
"age",
"education-num",
"hours-per-week",
]
kde_distributions(
df=df,
n_rows=1,
n_cols=3,
grid_figsize=(14, 4),
fill=True,
fill_alpha=0.60,
text_wrap=50,
bbox_inches="tight",
vars_of_interest=vars_of_interest,
y_axis_label="Density",
bins=10,
plot_type="both",
label_fontsize=16,
tick_fontsize=14,
)
Histogram Example (Density)
In this example, the kde_distributions() function is used to generate histograms for
the variables "age", "education-num", and "hours-per-week" but with
plot_type="hist", meaning no KDE plots are included—only histograms are displayed.
The plots are arranged in a single row of four columns (n_rows=1, n_cols=3),
with a grid size of 14x4 inches (grid_figsize=(14, 4)). The histograms are
divided into 10 bins (bins=10), and the y-axis is labeled “Density” (y_axis_label="Density").
Font sizes for the axis labels and tick labels are set to 16 and 14 points,
respectively, ensuring clarity in the visualizations. This setup focuses on the
histogram representation without the KDE overlay.
from eda_toolkit import kde_distributions
vars_of_interest = [
"age",
"education-num",
"hours-per-week",
]
kde_distributions(
df=df,
n_rows=1,
n_cols=3,
grid_figsize=(14, 4),
fill=True,
text_wrap=50,
bbox_inches="tight",
vars_of_interest=vars_of_interest,
y_axis_label="Density",
bins=10,
plot_type="hist",
label_fontsize=16,
tick_fontsize=14,
)
Histogram Example (Count)
In this example, the kde_distributions() function is modified to generate histograms
with a few key changes. The hist_color is set to “orange”, changing the color of the
histogram bars. The y-axis label is updated to “Count” (y_axis_label="Count"),
reflecting that the histograms display the count of observations within each bin.
Additionally, the stat parameter is set to "Count" to show the actual counts instead of
densities. The rest of the parameters remain the same as in the previous example,
with the plots arranged in a single row of four columns (n_rows=1, n_cols=3),
a grid size of 14x4 inches, and a bin count of 10. This setup focuses on
visualizing the raw counts in the dataset using orange-colored histograms.
from eda_toolkit import kde_distributions
vars_of_interest = [
"age",
"education-num",
"hours-per-week",
]
kde_distributions(
df=df,
n_rows=1,
n_cols=3,
grid_figsize=(14, 4),
text_wrap=50,
hist_color="orange",
bbox_inches="tight",
vars_of_interest=vars_of_interest,
y_axis_label="Count",
bins=10,
plot_type="hist",
stat="Count",
label_fontsize=16,
tick_fontsize=14,
)
Histogram Example - (Mean and Median)
In this example, the kde_distributions() function is customized to generate
histograms that include mean and median lines. The mean_color is set to "blue"
and the median_color is set to "black", allowing for a clear distinction
between the two statistical measures. The function parameters are adjusted to
ensure that both the mean and median lines are plotted (plot_mean=True, plot_median=True).
The y_axis_label remains "Density", indicating that the histograms
represent the density of observations within each bin. The histogram bars are
colored using hist_color="brown", with a fill_alpha=0.60 while the s
tatistical overlays enhance the interpretability of the data. The layout is
configured with a single row and multiple columns (n_rows=1, n_cols=3), and
the grid size is set to 15x5 inches. This example highlights how to visualize
central tendencies within the data using a histogram that prominently displays
the mean and median.
from eda_toolkit import kde_distributions
vars_of_interest = [
"age",
"education-num",
"hours-per-week",
]
kde_distributions(
df=df,
n_rows=1,
n_cols=3,
grid_figsize=(14, 4),
text_wrap=50,
hist_color="brown",
bbox_inches="tight",
vars_of_interest=vars_of_interest,
y_axis_label="Density",
bins=10,
fill_alpha=0.60,
plot_type="hist",
stat="Density",
label_fontsize=16,
tick_fontsize=14,
plot_mean=True,
plot_median=True,
mean_color="blue",
)
Histogram Example - (Mean, Median, and Std. Deviation)
In this example, the kde_distributions() function is customized to generate
a histogram that include mean, median, and 3 standard deviation lines. The
mean_color is set to "blue" and the median_color is set to "black",
allowing for a clear distinction between these two central tendency measures.
The function parameters are adjusted to ensure that both the mean and median lines
are plotted (plot_mean=True, plot_median=True). The y_axis_label remains
"Density", indicating that the histograms represent the density of observations
within each bin. The histogram bars are colored using hist_color="brown",
with a fill_alpha=0.40, which adjusts the transparency of the fill color.
Additionally, standard deviation bands are plotted using colors "purple",
"green", and "silver" for one, two, and three standard deviations, respectively.
The layout is configured with a single row and multiple columns (n_rows=1, n_cols=3),
and the grid size is set to 15x5 inches. This setup is particularly useful for
visualizing the central tendencies within the data while also providing a clear
view of the distribution and spread through the standard deviation bands. The
configuration used in this example showcases how histograms can be enhanced with
statistical overlays to provide deeper insights into the data.
Note
You have the freedom to choose whether to plot the mean, median, and standard deviation lines. You can display one, none, or all of these simultaneously.
from eda_toolkit import kde_distributions
vars_of_interest = [
"age",
]
kde_distributions(
df=df,
figsize=(10, 6),
text_wrap=50,
hist_color="brown",
bbox_inches="tight",
vars_of_interest=vars_of_interest,
y_axis_label="Density",
bins=10,
fill_alpha=0.40,
plot_type="both",
stat="Density",
label_fontsize=16,
tick_fontsize=14,
plot_mean=True,
plot_median=True,
mean_color="blue",
std_dev_levels=[
1,
2,
3,
],
std_color=[
"purple",
"green",
"silver",
],
)
Feature Scaling and Outliers
- data_doctor(df, feature_name, data_fraction=1, scale_conversion=None, scale_conversion_kws=None, apply_cutoff=False, lower_cutoff=None, upper_cutoff=None, show_plot=True, plot_type='all', figsize=(18, 6), xlim=None, kde_ylim=None, hist_ylim=None, box_violin_ylim=None, save_plot=False, image_path_png=None, image_path_svg=None, apply_as_new_col_to_df=False, kde_kws=None, hist_kws=None, box_violin_kws=None, box_violin='boxplot', label_fontsize=12, tick_fontsize=10, random_state=None)
Analyze and transform a specific feature in a DataFrame, with options for scaling, applying cutoffs, and visualizing the results. This function also allows for the creation of a new column with the transformed data if specified. Plots can be saved in PNG or SVG format with filenames that incorporate the
plot_type,feature_name,scale_conversion, andcutoffif cutoffs are applied.- Parameters:
df (pandas.DataFrame) – The DataFrame containing the feature to analyze.
feature_name (str) – The name of the feature (column) to analyze.
data_fraction (float, optional) – Fraction of the data to analyze. Default is
1(full dataset). Useful for large datasets where a sample can represent the population. Ifapply_as_new_col_to_df=True, the full dataset is used (data_fraction=1).scale_conversion (str, optional) –
Type of conversion to apply to the feature. Options include:
'abs': Absolute values'log': Natural logarithm'sqrt': Square root'cbrt': Cube root'reciprocal': Reciprocal transformation'stdrz': Standardized (z-score)'minmax': Min-Max scaling'boxcox': Box-Cox transformation (positive values only; supportslmbdafor specific lambda oralphafor confidence interval)'robust': Robust scaling (median and IQR)'maxabs': Max-abs scaling'exp': Exponential transformation'logit': Logit transformation (values between 0 and 1)'arcsinh': Inverse hyperbolic sine'square': Squaring the values'power': Power transformation (Yeo-Johnson).
scale_conversion_kws (dict, optional) –
Additional keyword arguments to pass to the scaling functions, such as:
'alpha'for Box-Cox transformation (returns a confidence interval for lambda)'lmbda'for a specific Box-Cox transformation value'quantile_range'for robust scaling.
apply_cutoff (bool, optional (default=False)) – Whether to apply upper and/or lower cutoffs to the feature.
lower_cutoff (float, optional) – Lower bound to apply if
apply_cutoff=True.upper_cutoff (float, optional) – Upper bound to apply if
apply_cutoff=True.show_plot (bool, optional (default=True)) – Whether to display plots of the transformed feature: KDE, histogram, and boxplot/violinplot.
plot_type (str, list, or tuple, optional (default="all")) –
Specifies the type of plot(s) to produce. Options are:
'all': Generates KDE, histogram, and boxplot/violinplot.'kde': KDE plot only.'hist': Histogram plot only.'box_violin': Boxplot or violin plot only (specified bybox_violin).
If a list or tuple is provided (e.g.,
plot_type=["kde", "hist"]), the specified plots are displayed in a single row with sufficient spacing. AValueErroris raised if an invalid plot type is included.figsize (tuple or list, optional (default=(18, 6))) – Specifies the figure size for the plots. This applies to all plot types, including single plots (when
plot_typeis set to “kde”, “hist”, or “box_violin”) and multi-plot layout whenplot_typeis “all”.xlim (tuple or list, optional) – Limits for the x-axis in all plots, specified as
(xmin, xmax).kde_ylim (tuple or list, optional) – Limits for the y-axis in the KDE plot, specified as
(ymin, ymax).hist_ylim (tuple or list, optional) – Limits for the y-axis in the histogram plot, specified as
(ymin, ymax).box_violin_ylim (tuple or list, optional) – Limits for the y-axis in the boxplot or violin plot, specified as
(ymin, ymax).save_plot (bool, optional (default=False)) – Whether to save the plots as PNG and/or SVG images. If
True, the user must specify at least one ofimage_path_pngorimage_path_svg, otherwise aValueErroris raised.image_path_png (str, optional) – Directory path to save the plot as a PNG file. Only used if
save_plot=True.image_path_svg (str, optional) – Directory path to save the plot as an SVG file. Only used if
save_plot=True.apply_as_new_col_to_df (bool, optional (default=False)) –
Whether to create a new column in the DataFrame with the transformed values. If
True, the new column name is generated based on the feature name and the transformation applied:<feature_name>_<scale_conversion>: If a transformation is applied.<feature_name>_w_cutoff: If only cutoffs are applied.
For Box-Cox transformation, if
alphais specified, the confidence interval for lambda is displayed. Iflmbdais specified, the lambda value is displayed.kde_kws (dict, optional) – Additional keyword arguments to pass to the KDE plot (
seaborn.kdeplot).hist_kws (dict, optional) – Additional keyword arguments to pass to the histogram plot (
seaborn.histplot).box_violin_kws (dict, optional) – Additional keyword arguments to pass to either boxplot or violinplot.
box_violin (str, optional (default="boxplot")) – Specifies whether to plot a
boxplotorviolinplotifplot_typeis set tobox_violin.label_fontsize (int, optional (default=12)) – Font size for the axis labels and plot titles.
tick_fontsize (int, optional (default=10)) – Font size for the tick labels on both axes.
random_state (int, optional) – Seed for reproducibility when sampling the data.
- Returns:
NoneDisplays the feature’s descriptive statistics, quartile information, and outlier details. If a new column is created, confirms the addition to the DataFrame. For Box-Cox, either the lambda or its confidence interval is displayed.- Raises:
If an invalid
scale_conversionis provided.If Box-Cox transformation is applied to non-positive values.
If
save_plot=Truebut neitherimage_path_pngnorimage_path_svgis provided.If an invalid option is provided for
box_violin.If an invalid option is provided for
plot_type.If the length of transformed data does not match the original feature length.
Note
When saving plots, the filename will include the
feature_name,scale_conversion, each selectedplot_type, and, if cutoffs are applied,"_cutoff". For example, iffeature_nameis"age",scale_conversionis"boxcox", andplot_typeis"kde", with cutoffs applied, the filename will be:age_boxcox_kde_cutoff.pngorage_boxcox_kde_cutoff.svg.
Available Scale Conversions
The scale_conversion parameter accepts several options for data scaling, providing flexibility in how you preprocess your data. Each option addresses specific transformation needs, such as normalizing data, stabilizing variance, or adjusting data ranges. Below is the exhaustive list of available scale conversions:
'abs': Takes the absolute values of the data, removing any negative signs.'log': Applies the natural logarithm to the data, useful for compressing large ranges and reducing skewness.'sqrt': Applies the square root transformation, often used to stabilize variance.'cbrt': Takes the cube root of the data, which can be useful for transforming both positive and negative values symmetrically.'stdrz': Standardizes the data to have a mean of 0 and a standard deviation of 1, also known as z-score normalization.'minmax': Rescales the data to a specified range, defaulting to [0, 1], ensuring that all values fall within this range.'boxcox': Applies the Box-Cox transformation to stabilize variance and make the data more normally distributed. Only works with positive values and supports passinglmbdaoralphafor flexibility.'robust': Scales the data based on percentiles (such as the interquartile range), which reduces the influence of outliers.'maxabs': Scales the data by dividing it by its maximum absolute value, preserving the sign of the data while constraining it to the range [-1, 1].'reciprocal': Transforms the data by taking the reciprocal (1/x), which is useful when handling values that are far from zero.'exp': Applies the exponential function to the data, which is useful for modeling exponential growth or increasing the impact of large values.'logit': Applies the logit transformation to data, which is only valid for values between 0 and 1. This is typically used in logistic regression models.'arcsinh': Applies the inverse hyperbolic sine transformation, which is similar to the logarithm but can handle both positive and negative values.'square': Squares the values of the data, effectively emphasizing larger values while downplaying smaller ones.'power': Applies the power transformation (Yeo-Johnson), which is similar to Box-Cox but works for both positive and negative values.
boxcox is just one of the many options available for transforming data in the data_doctor function, providing versatility to handle different scaling needs.
Box-Cox Transformation Example 1
In this example from the US Census dataset [1], we demonstrate the usage of the data_doctor
function to apply a Box-Cox transformation to the age column in a DataFrame.
The data_doctor function provides a flexible way to preprocess data by applying
various scaling techniques. In this case, we apply the Box-Cox transformation without any tuning
of the alpha or lambda parameters, allowing the function to handle the transformation in a
barebones approach. You can also choose other scaling conversions from the list of available
options (such as 'minmax', 'standard', 'robust', etc.), depending on your needs.
from eda_toolkit import data_doctor
data_doctor(
df=df,
feature_name="age",
data_fraction=0.6,
scale_conversion="boxcox",
apply_cutoff=False,
lower_cutoff=None,
upper_cutoff=None,
show_plot=True,
apply_as_new_col_to_df=True,
random_state=111,
)
DATA DOCTOR SUMMARY REPORT
+------------------------------+--------------------+
| Feature | age |
+------------------------------+--------------------+
| Statistic | Value |
+------------------------------+--------------------+
| Min | 3.6664 |
| Max | 6.8409 |
| Mean | 5.0163 |
| Median | 5.0333 |
| Std Dev | 0.6761 |
+------------------------------+--------------------+
| Quartile | Value |
+------------------------------+--------------------+
| Q1 (25%) | 4.5219 |
| Q2 (50% = Median) | 5.0333 |
| Q3 (75%) | 5.5338 |
| IQR | 1.0119 |
+------------------------------+--------------------+
| Outlier Bound | Value |
+------------------------------+--------------------+
| Lower Bound | 3.0040 |
| Upper Bound | 7.0517 |
+------------------------------+--------------------+
New Column Name: age_boxcox
Box-Cox Lambda: 0.1748
df.head()
| age | workclass | education | education-num | marital-status | occupation | relationship | age_boxcox | |
|---|---|---|---|---|---|---|---|---|
| census_id | ||||||||
| 582248222 | 39 | State-gov | Bachelors | 13 | Never-married | Adm-clerical | Not-in-family | 5.180807 |
| 561810758 | 50 | Self-emp-not-inc | Bachelors | 13 | Married-civ-spouse | Exec-managerial | Husband | 5.912323 |
| 598098459 | 38 | Private | HS-grad | 9 | Divorced | Handlers-cleaners | Not-in-family | 5.227960 |
| 776705221 | 53 | Private | 11th | 7 | Married-civ-spouse | Handlers-cleaners | Husband | 6.389562 |
| 479262902 | 28 | Private | Bachelors | 13 | Married-civ-spouse | Prof-specialty | Wife | 3.850675 |
Note
Notice that the unique identifiers function was also applied on the dataframe to generate randomized census IDs for the rows of the data.
Explanation
df=df: We are passingdfas the input DataFrame.feature_name="age": The feature we are transforming isage.data_fraction=1: We are using 100% of the data in theagecolumn. You can adjust this if you want to perform the operation on a subset of the data.scale_conversion="boxcox": This parameter defines the type of scaling we want to apply. In this case, we are using the Box-Cox transformation. You can changeboxcoxto any supported scale conversion method.apply_cutoff=False: We are not applying any outlier cutoff in this example.lower_cutoff=Noneandupper_cutoff=None: These are left asNonesince we are not applying outlier cutoffs in this case.show_plot=True: This option will generate a plot to visualize the distribution of theagecolumn before and after the transformation.apply_as_new_col_to_df=True: This tells the function to apply the transformation and create a new column in the DataFrame. The new column will be namedage_boxcox, where"boxcox"indicates the type of transformation applied.
Box-Cox Transformation: This transformation normalizes the data by making the distribution more Gaussian-like, which can be beneficial for certain statistical models.
No Outlier Handling: In this example, we are not applying any cutoffs to remove or modify outliers. This means the function will process the entire range of values in the
agecolumn without making adjustments for extreme values.New Column Creation: By setting
apply_as_new_col_to_df=True, a new column namedage_boxcoxwill be created in thedfDataFrame, where the transformed values will be stored. This allows us to keep the originalagecolumn intact while adding the transformed data as a new feature.The
show_plot=Trueparameter will generate a plot that visualizes the distribution of the originalagedata alongside the transformedage_boxcoxdata. This can help you assess how the Box-Cox transformation has affected the data distribution.
Box-Cox Transformation Example 2
In this second example from the US Census dataset [1], we apply the Box-Cox
transformation to the age column in a DataFrame, but this time with custom
keyword arguments passed through the scale_conversion_kws. Specifically, we
provide an alpha value of 0.8, influencing the confidence interval for the
transformation. Additionally, we customize the
visual appearance of the plots by specifying keyword arguments for the violinplot,
KDE, and histogram plots. These customizations allow for greater control over the
visual output.
from eda_toolkit import data_doctor
data_doctor(
df=df,
feature_name="age",
data_fraction=1,
scale_conversion="boxcox",
apply_cutoff=False,
lower_cutoff=None,
upper_cutoff=None,
show_plot=True,
apply_as_new_col_to_df=True,
scale_conversion_kws={"alpha": 0.8},
box_violin="violinplot",
box_violin_kws={"color": "lightblue"},
kde_kws={"fill": True, "color": "blue"},
hist_kws={"color": "green"},
random_state=111,
)
DATA DOCTOR SUMMARY REPORT
+------------------------------+--------------------+
| Feature | age |
+------------------------------+--------------------+
| Statistic | Value |
+------------------------------+--------------------+
| Min | 3.6664 |
| Max | 6.8409 |
| Mean | 5.0163 |
| Median | 5.0333 |
| Std Dev | 0.6761 |
+------------------------------+--------------------+
| Quartile | Value |
+------------------------------+--------------------+
| Q1 (25%) | 4.5219 |
| Q2 (50% = Median) | 5.0333 |
| Q3 (75%) | 5.5338 |
| IQR | 1.0119 |
+------------------------------+--------------------+
| Outlier Bound | Value |
+------------------------------+--------------------+
| Lower Bound | 3.0040 |
| Upper Bound | 7.0517 |
+------------------------------+--------------------+
New Column Name: age_boxcox
Box-Cox C.I. for Lambda: (0.1717, 0.1779)
Note
Note that this example specifies The theoretical overview section provides a detailed framework for a Box-Cox transformation.
df.head()
| age | workclass | education | education-num | marital-status | occupation | relationship | age_boxcox | |
|---|---|---|---|---|---|---|---|---|
| census_id | ||||||||
| 582248222 | 39 | State-gov | Bachelors | 13 | Never-married | Adm-clerical | Not-in-family | 3.936876 |
| 561810758 | 50 | Self-emp-not-inc | Bachelors | 13 | Married-civ-spouse | Exec-managerial | Husband | 4.019590 |
| 598098459 | 38 | Private | HS-grad | 9 | Divorced | Handlers-cleaners | Not-in-family | 4.521908 |
| 776705221 | 53 | Private | 11th | 7 | Married-civ-spouse | Handlers-cleaners | Husband | 5.033257 |
| 479262902 | 28 | Private | Bachelors | 13 | Married-civ-spouse | Prof-specialty | Wife | 5.614411 |
In this example, you can see how the data_doctor function supports further
flexibility with customizable plot aesthetics and scaling techniques. The
Box-Cox transformation is still applied without any tuning of the lambda parameter,
while the alpha value provides a confidence interval for the resulting transformation:
Box-Cox C.I. for Lambda: (0.1717, 0.1779)
This allows for tailored visualizations with consistent styling across multiple plot types.
Some of the keyword arguments, such as those passed in box_violin_kws, are
specific to Python version 3.7. For example, in this version, we remove the fill
color from the boxplot using boxprops.
box_violin_kws={
"boxprops": dict(facecolor="none", edgecolor="blue")
},
In later Python versions (e.g., 3.11),
this can be done more easily with fill=True. Therefore, it is important to pass
any desired keyword arguments based on the correct version of Python you’re using.
Explanation
df=df: We are passingdfas the input DataFrame.feature_name="age": The feature we are transforming isage.data_fraction=1: We are using 100% of the data in theagecolumn. You can adjust this if you want to perform the operation on a subset of the data.scale_conversion="boxcox": This parameter defines the type of scaling we want to apply. In this case, we are using the Box-Cox transformation.apply_cutoff=False: We are not applying any outlier cutoff in this example.lower_cutoff=Noneandupper_cutoff=None: These are left asNonesince we are not applying outlier cutoffs in this case.show_plot=True: This option will generate a plot to visualize the distribution of theagecolumn before and after the transformation.apply_as_new_col_to_df=True: This tells the function to apply the transformation and create a new column in the DataFrame. The new column will be namedage_boxcox_alphato indicate that an alpha parameter was used in the transformation.scale_conversion_kws={"alpha":0.8}: Thealphakeyword argument specifies the confidence interval for the Box-Cox transformation’s lambda value, ensuring a confidence interval is returned instead of a single lambda value.box_violin_kws={"boxprops": dict(facecolor='none', edgecolor="blue")}: This keyword argument customizes the appearance of the boxplot by removing the fill color and setting the edge color to blue. This syntax is specific to Python 3.7. In later versions (i.e., 3.11+), thefill=Trueargument can be used to control this behavior.kde_kws={"fill":True, "color":"blue"}: This fills the area under the KDE plot with a blue color, enhancing the plot’s visual presentation.hist_kws={"color":"blue"}: This colors the histogram bars in blue for visual consistency across plots.image_path_svg=image_path_svg: This parameter specifies the path where the resulting plot will be saved as an SVG file.save_plot=True: This tells the function to save the plot, and since an image path is provided, the plot will be saved as an SVG file.
Box-Cox Transformation with Confidence Interval: In this example, we use the Box-Cox transformation with the
alphaparameter set to 0.8, which returns a confidence interval for the lambda value rather than a single value.No Outlier Handling: Similar to Example 1, no outliers are handled in this transformation.
New Column Creation: The transformed data is added to the DataFrame in a new column named
age_boxcox_alpha, where “alpha” indicates the confidence interval applied in the Box-Cox transformation.Custom Plot Visuals: The KDE, histogram, and boxplot are customized with blue colors, and specific keyword arguments are provided for the boxplot appearance based on Python version. These changes allow for finer control over the visual aesthetics of the resulting plots.
Plot Saving: The
save_plotparameter is set toTrue, and the plot will be saved as an SVG file at the specified location.
Data Fraction Usage
In the Box-Cox transformation examples, you may notice a difference in the values for data_fraction:
In Box-Cox Example 1, we set
data_fraction=0.6.In Box-Cox Example 2, we used the full data with
data_fraction=1.
Despite using a data_fraction of 0.6 in Example 1, the function still processed
the entire dataset. The purpose of the data_fraction parameter is to allow
users to select a smaller subset of the data for sampling and transformation while
ensuring the final operation is applied to the full scope of data.
This behavior is intentional, as it serves to:
1. Ensure Reproducibility: By using a consistent random_state, the sampled
subset can reliably represent the dataset, regardless of data_fraction.
2. Preserve Sampling Assumptions: Applying the desired operation (e.g., transformations) on the full data aligns the sample with the larger population and allows a seamless projection of the sample properties to the entire dataset.
Thus, while data_fraction provides a way to adjust the percentage of data
used for sampling, the function will always apply the transformation across the
full dataset, balancing performance efficiency with statistical integrity.
Retaining a Sample for Analysis
To sample the exact subset used in the data_fraction=0.6 calculation, you
can directly sample from the DataFrame with a consistent random state for
reproducibility. This method allows you to work with a representative subset of
the data while preserving the original distribution characteristics.
To sample 60% of the data using the exact logic of the data_doctor function,
use the following code:
sampled_df = df.sample(frac=0.6, random_state=111)
The random_state parameter ensures that the sampled data remains consistent
across runs. After creating this subset, you can apply the data_doctor
function to sampled_df as shown below to perform the Box-Cox transformation
on the age column:
from eda_toolkit import data_doctor
data_doctor(
df=sampled_df,
feature_name="age",
data_fraction=1,
scale_conversion="boxcox",
apply_cutoff=False,
lower_cutoff=None,
upper_cutoff=None,
show_plot=True,
apply_as_new_col_to_df=True,
random_state=111,
)
By setting data_fraction=1 within the data_doctor function, you ensure
that it operates on the entire sampled_df, which now consists of the selected
60% subset. To confirm that the sampled data is indeed 60% of the original
DataFrame, you can print the shape of sampled_df as follows:
print(
f"The sampled dataframe has {sampled_df.shape[0]} rows and {sampled_df.shape[1]} columns."
)
The sampled dataframe has 29305 rows and 16 columns.
We can also inspect the first five rows of the sampled_df dataframe below:
| age | workclass | education | education-num | marital-status | occupation | relationship | age_boxcox | |
|---|---|---|---|---|---|---|---|---|
| census_id | ||||||||
| 408117383 | 40 | Private | Some-college | 10 | Married-civ-spouse | Machine-op-inspct | Husband | 4.355015 |
| 669717925 | 58 | Private | HS-grad | 9 | Married-civ-spouse | Exec-managerial | Husband | 5.086108 |
| 399428377 | 41 | Private | HS-grad | 9 | Separated | Machine-op-inspct | Not-in-family | 5.037743 |
| 961427355 | 73 | NaN | Some-college | 10 | Married-civ-spouse | NaN | Husband | 4.216561 |
| 458295720 | 19 | Private | HS-grad | 9 | Never-married | Farming-fishing | Not-in-family | 5.520438 |
Logit Transformation Example
In this example, we demonstrate the usage of the data_doctor function to
apply a logit transformation to a feature in a DataFrame. The logit transformation
is used when dealing with data bounded between 0 and 1, as it maps values within
this range to an unbounded scale in log-odds terms, making it particularly useful
in fields such as logistic regression.
Note
The data_doctor function provides a range of scaling options, and in this case,
we use the logit transformation to illustrate how the transformation is applied.
However, it’s important to note that if the feature contains values outside the (0, 1)
range, the function will raise a ValueError. This is because the logit function
is undefined for values less than or equal to 0 and greater than or equal to 1.
from eda_toolkit import data_doctor
data_doctor(
df=df,
feature_name="age",
data_fraction=1,
scale_conversion="logit",
apply_cutoff=False,
lower_cutoff=None,
upper_cutoff=None,
show_plot=True,
apply_as_new_col_to_df=True,
random_state=111,
)
Error
ValueError: Logit transformation requires values to be between 0 and 1. Consider using a scaling method such as min-max scaling first.
If you attempt to apply this transformation to data outside the (0, 1) range, such as an unscaled numerical feature, the function will halt and display an error message advising you to use an appropriate scaling method first.
If you encounter this error, it is recommended to first scale your data using a method like min-max scaling to bring it within the (0, 1) range before applying the logit transformation.
In this example:
df=df: Specifies the DataFrame containing the feature.feature_name="feature_proportion": The feature we are transforming should be bounded between 0 and 1.scale_conversion="logit": Sets the transformation to logit. Ensure thatfeature_proportionvalues are within (0, 1) before applying.show_plot=True: Generates a plot of the transformed feature.
Plain Outliers Example
Observed Outliers Sans Cutoffs
In this example, we examine the final weight (fnlwgt) feature from the US Census
dataset [1], focusing on detecting outliers without applying any scaling
transformations. The data_doctor function is used with minimal configuration
to visualize where outliers are present in the raw data.
By enabling apply_cutoff=True and selecting plot_type=["box_violin", "hist"],
we can clearly identify outliers both visually and numerically. This basic setup
highlights the outliers without altering the data distribution, making it easy
to see extreme values that could affect further analysis.
The following code demonstrates this:
from eda_toolkit import data_doctor
data_doctor(
df=df,
feature_name="fnlwgt",
data_fraction=0.6,
plot_type=["box_violin", "hist"],
hist_kws={"color": "gray"},
figsize=(8, 4),
image_path_svg=image_path_svg,
save_plot=True,
random_state=111,
)
DATA DOCTOR SUMMARY REPORT
+------------------------------+--------------------+
| Feature | fnlwgt |
+------------------------------+--------------------+
| Statistic | Value |
+------------------------------+--------------------+
| Min | 12,285.0000 |
| Max | 1,484,705.0000 |
| Mean | 189,181.3719 |
| Median | 177,955.0000 |
| Std Dev | 105,417.5713 |
+------------------------------+--------------------+
| Quartile | Value |
+------------------------------+--------------------+
| Q1 (25%) | 117,292.0000 |
| Q2 (50% = Median) | 177,955.0000 |
| Q3 (75%) | 236,769.0000 |
| IQR | 119,477.0000 |
+------------------------------+--------------------+
| Outlier Bound | Value |
+------------------------------+--------------------+
| Lower Bound | -61,923.5000 |
| Upper Bound | 415,984.5000 |
+------------------------------+--------------------+
In this visualization, the boxplot and histogram display outliers prominently, showing you exactly where the extreme values lie. This setup serves as a baseline view of the raw data, making it useful for assessing the initial distribution before any scaling or transformation is applied.
Treated Outliers With Cutoffs
In this scenario, we address the extreme values observed in the fnlwgt feature
by applying a visual cutoff based on the distribution seen in the previous example.
Here, we set an approximate upper cutoff of 400,000 to limit the impact of outliers
without any additional scaling or transformation. By using apply_cutoff=True along
with upper_cutoff=400000, we effectively cap the extreme values.
This example also demonstrates how you can further customize the visualization by
specifying additional histogram keyword arguments with hist_kws. Here, we use
bins=20 to adjust the bin size, creating a smoother view of the feature’s
distribution within the cutoff limits.
In the resulting visualization, you will see that the boxplot and histogram have a controlled range due to the applied upper cutoff, limiting the influence of extreme outliers on the visual representation. This treatment provides a clearer view of the primary distribution, allowing for a more focused analysis on the bulk of the data without outliers distorting the scale.
The following code demonstrates this configuration:
from eda_toolkit import data_doctor
data_doctor(
df=df,
feature_name="fnlwgt",
data_fraction=0.6,
apply_as_new_col_to_df=True,
apply_cutoff=True,
upper_cutoff=400000,
plot_type=["box_violin", "hist"],
hist_kws={"color": "gray", "bins": 20},
figsize=(8, 4),
image_path_svg=image_path_svg,
save_plot=True,
random_state=111,
)
| age | workclass | fnlwgt | education | marital-status | occupation | relationship | fnlwgt_w_cutoff | |
|---|---|---|---|---|---|---|---|---|
| census_id | ||||||||
| 582248222 | 39 | State-gov | 77516 | Bachelors | Never-married | Adm-clerical | Not-in-family | 132222 |
| 561810758 | 50 | Self-emp-not-inc | 83311 | Bachelors | Married-civ-spouse | Exec-managerial | Husband | 68624 |
| 598098459 | 38 | Private | 215646 | HS-grad | Divorced | Handlers-cleaners | Not-in-family | 161880 |
| 776705221 | 53 | Private | 234721 | 11th | Married-civ-spouse | Handlers-cleaners | Husband | 73402 |
| 479262902 | 28 | Private | 338409 | Bachelors | Married-civ-spouse | Prof-specialty | Wife | 97261 |
RobustScaler Outliers Examples
In this example from the US Census dataset [1], we apply the RobustScaler
transformation to the age column in a DataFrame to address potential outliers.
The data_doctor function enables users to apply transformations with specific
configurations via the scale_conversion_kws parameter, making it ideal for
refining how outliers affect scaling.
For this example, we set the following custom keyword arguments:
Disable centering: By setting
with_centering=False, the transformation scales based only on the range, without shifting the median to zero.Adjust quantile range: We specify a narrower
quantile_rangeof (10.0, 90.0) to reduce the influence of extreme values on scaling.
The following code demonstrates this transformation:
from eda_toolkit import data_doctor
data_doctor(
df=df,
feature_name='age',
data_fraction=0.6,
scale_conversion="robust",
apply_as_new_col_to_df=True,
scale_conversion_kws={
"with_centering": False, # Disable centering
"quantile_range": (10.0, 90.0) # Use a custom quantile range
},
random_state=111,
)
DATA DOCTOR SUMMARY REPORT
+------------------------------+--------------------+
| Feature | age |
+------------------------------+--------------------+
| Statistic | Value |
+------------------------------+--------------------+
| Min | 0.4722 |
| Max | 2.5000 |
| Mean | 1.0724 |
| Median | 1.0278 |
| Std Dev | 0.3809 |
+------------------------------+--------------------+
| Quartile | Value |
+------------------------------+--------------------+
| Q1 (25%) | 0.7778 |
| Q2 (Median) | 1.0278 |
| IQR | 0.5556 |
| Q3 (75%) | 1.3333 |
| Q4 (Max) | 2.5000 |
+------------------------------+--------------------+
| Outlier Bound | Value |
+------------------------------+--------------------+
| Lower Bound | -0.0556 |
| Upper Bound | 2.1667 |
+------------------------------+--------------------+
New Column Name: age_robust
Stacked Crosstab Plots
Generate stacked or regular bar plots and crosstabs for specified columns in a DataFrame.
The stacked_crosstab_plot function is a powerful tool for visualizing categorical data relationships through stacked bar plots and contingency tables (crosstabs). It supports extensive customization options, including plot appearance, color schemes, and saving output in multiple formats. Users can choose between regular or normalized plots and control whether the function returns the generated crosstabs as a dictionary.
- stacked_crosstab_plot(df, col, func_col, legend_labels_list, title, kind='bar', width=0.9, rot=0, custom_order=None, image_path_png=None, image_path_svg=None, save_formats=None, color=None, output='both', return_dict=False, x=None, y=None, p=None, file_prefix=None, logscale=False, plot_type='both', show_legend=True, label_fontsize=12, tick_fontsize=10, text_wrap=50, remove_stacks=False, xlim=None, ylim=None)
- Parameters:
df (pandas.DataFrame) – The DataFrame containing the data to plot.
col (str) – The name of the column in the DataFrame to be analyzed.
func_col (list of str) – List of columns in the DataFrame to generate the crosstabs and stack the bars in the plot.
legend_labels_list (list of list of str) – List of legend labels corresponding to each column in
func_col.title (list of str) – List of titles for each plot generated.
kind (str, optional) – Type of plot to generate (
"bar"or"barh"for horizontal bars). Default is"bar".width (float, optional) – Width of the bars in the bar plot. Default is
0.9.rot (int, optional) – Rotation angle of the x-axis labels. Default is
0.custom_order (list, optional) – Custom order for the categories in
col.image_path_png (str, optional) – Directory path to save PNG plot images.
image_path_svg (str, optional) – Directory path to save SVG plot images.
save_formats (list, str, or tuple, optional) – List, string, or tuple of file formats to save the plots (e.g.,
["png", "svg"]). Default isNone. Raises an error if an invalid format is provided without the corresponding path.color (list of str, optional) – List of colors to use for the plots. Default is the seaborn color palette.
output (str, optional) – Specify the output type:
"plots_only","crosstabs_only", or"both". Default is"both".return_dict (bool, optional) – Return the crosstabs as a dictionary. Default is
False.x (int, optional) – Width of the figure in inches.
y (int, optional) – Height of the figure in inches.
p (int, optional) – Padding between subplots.
file_prefix (str, optional) – Prefix for filenames when saving plots.
logscale (bool, optional) – Apply a logarithmic scale to the y-axis. Default is
False.plot_type (str, optional) – Type of plot to generate:
"both","regular", or"normalized". Default is"both".show_legend (bool, optional) – Show the legend on the plot. Default is
True.label_fontsize (int, optional) – Font size for axis labels. Default is
12.tick_fontsize (int, optional) – Font size for tick labels. Default is
10.text_wrap (int, optional) – Maximum width of the title text before wrapping. Default is
50.remove_stacks (bool, optional) – Remove stacks and create a regular bar plot. Only works when
plot_typeis"regular". Default isFalse.xlim (tuple or list, optional) – Tuple or list specifying limits of the x-axis (e.g.,
(min, max)).ylim (tuple or list, optional) – Tuple or list specifying limits of the y-axis (e.g.,
(min, max)).
- Raises:
If
remove_stacksisTrueandplot_typeis not"regular".If an invalid save format is specified without providing the corresponding path.
If
outputis not one of"both","plots_only", or"crosstabs_only".If
plot_typeis not one of"both","regular", or"normalized".If
remove_stacksis used withplot_typenot set to"regular".If lengths of
title,func_col, andlegend_labels_listare unequal.
KeyError – If any column in
colorfunc_colis missing from the DataFrame.
- Returns:
Dictionary of crosstabs DataFrames if
return_dictisTrue. Otherwise, returnsNone.- Return type:
dict or None
Notes
Ensure
save_formatsaligns with provided paths for saving images. For instance, specifyimage_path_pngorimage_path_svgif saving as"png"or"svg"respectively.The returned crosstabs dictionary includes both absolute and normalized values when
return_dict=True.
Stacked Bar Plots With Crosstabs Example
The provided code snippet demonstrates how to use the stacked_crosstab_plot
function to generate stacked bar plots and corresponding crosstabs for different
columns in a DataFrame. Here’s a detailed breakdown of the code using the census
dataset as an example [1].
First, the func_col list is defined, specifying the columns ["sex", "income"]
to be analyzed. These columns will be used in the loop to generate separate plots.
The legend_labels_list is then defined, with each entry corresponding to a
column in func_col. In this case, the labels for the sex column are
["Male", "Female"], and for the income column, they are ["<=50K", ">50K"].
These labels will be used to annotate the legends of the plots.
Next, the title list is defined, providing titles for each plot corresponding
to the columns in func_col. The titles are set to ["Sex", "Income"],
which will be displayed on top of each respective plot.
Note
The legend_labels_list parameter should be a list of lists, where each
inner list corresponds to the ground truth labels for the respective item in
the func_col list. Each element in the func_col list represents a
column in your DataFrame that you wish to analyze, and the corresponding
inner list in legend_labels_list should contain the labels that will be
used in the legend of your plots.
For example:
# Define the func_col to use in the loop in order of usage
func_col = ["sex", "income"]
# Define the legend_labels to use in the loop
legend_labels_list = [
["Male", "Female"], # Corresponds to "sex"
["<=50K", ">50K"], # Corresponds to "income"
]
# Define titles for the plots
title = [
"Sex",
"Income",
]
Important
Ensure that func_col, legend_labels_list, and title have the
same number of elements. Each item in func_col must correspond to a list
of labels in legend_labels_list and a title in title to ensure the
function generates plots with the correct labels and titles.
Additionally, in this example, remove trailing periods from the income
column to correctly split its contents into two categories.
In this example:
func_colcontains two elements:"sex"and"income". Each corresponds to a specific column in your DataFrame.legend_labels_listis a nested list containing two inner lists:The first inner list,
["Male", "Female"], corresponds to the"sex"column infunc_col.The second inner list,
["<=50K", ">50K"], corresponds to the"income"column infunc_col.
titlecontains two elements:"Sex"and"Income", which will be used as the titles for the respective plots.
Note
Before proceeding with any further examples in this documentation, ensure that the age variable is binned into a new variable, age_group.
Detailed instructions for this process can be found under Binning Numerical Columns.
from eda_toolkit import stacked_crosstab_plot
stacked_crosstabs = stacked_crosstab_plot(
df=df,
col="age_group",
func_col=func_col,
legend_labels_list=legend_labels_list,
title=title,
kind="bar",
width=0.8,
rot=0,
custom_order=None,
color=["#00BFC4", "#F8766D"],
output="both",
return_dict=True,
x=14,
y=8,
p=10,
logscale=False,
plot_type="both",
show_legend=True,
label_fontsize=14,
tick_fontsize=12,
)
The above example generates stacked bar plots for "sex" and "income"
grouped by "education". The plots are executed with legends, labels, and
tick sizes customized for clarity. The function returns a dictionary of
crosstabs for further analysis or export.
Important
Importance of Correctly Aligning Labels
It is crucial to properly align the elements in the legend_labels_list,
title, and func_col parameters when using the stacked_crosstab_plot
function. Each of these lists must be ordered consistently because the function
relies on their alignment to correctly assign labels and titles to the
corresponding plots and legends.
For instance, in the example above:
The first element in
func_colis"sex", and it is aligned with the first set of labels["Male", "Female"]inlegend_labels_listand the first title"Sex"in thetitlelist.Similarly, the second element in
func_col,"income", aligns with the labels["<=50K", ">50K"]and the title"Income".
Misalignment between these lists would result in incorrect labels or titles being applied to the plots, potentially leading to confusion or misinterpretation of the data. Therefore, it’s important to ensure that each list is ordered appropriately and consistently to accurately reflect the data being visualized.
Proper Setup of Lists
When setting up the legend_labels_list, title, and func_col, ensure
that each element in the lists corresponds to the correct variable in the DataFrame.
This involves:
Ordering: Maintaining the same order across all three lists to ensure that labels and titles correspond correctly to the data being plotted.
Consistency: Double-checking that each label in
legend_labels_listmatches the categories present in the correspondingfunc_col, and that thetitleaccurately describes the plot.
By adhering to these guidelines, you can ensure that the stacked_crosstab_plot
function produces accurate and meaningful visualizations that are easy to interpret and analyze.
Output
Note
When you set return_dict=True, you are able to see the crosstabs printed out
as shown below.
| Crosstab for sex | |||||
|---|---|---|---|---|---|
| sex | Female | Male | Total | Female_% | Male_% |
| age_group | |||||
| < 18 | 295 | 300 | 595 | 49.58 | 50.42 |
| 18-29 | 5707 | 8213 | 13920 | 41 | 59 |
| 30-39 | 3853 | 9076 | 12929 | 29.8 | 70.2 |
| 40-49 | 3188 | 7536 | 10724 | 29.73 | 70.27 |
| 50-59 | 1873 | 4746 | 6619 | 28.3 | 71.7 |
| 60-69 | 939 | 2115 | 3054 | 30.75 | 69.25 |
| 70-79 | 280 | 535 | 815 | 34.36 | 65.64 |
| 80-89 | 40 | 91 | 131 | 30.53 | 69.47 |
| 90-99 | 17 | 38 | 55 | 30.91 | 69.09 |
| Total | 16192 | 32650 | 48842 | 33.15 | 66.85 |
| Crosstab for income | |||||
| income | <=50K | >50K | Total | <=50K_% | >50K_% |
| age_group | |||||
| < 18 | 595 | 0 | 595 | 100 | 0 |
| 18-29 | 13174 | 746 | 13920 | 94.64 | 5.36 |
| 30-39 | 9468 | 3461 | 12929 | 73.23 | 26.77 |
| 40-49 | 6738 | 3986 | 10724 | 62.83 | 37.17 |
| 50-59 | 4110 | 2509 | 6619 | 62.09 | 37.91 |
| 60-69 | 2245 | 809 | 3054 | 73.51 | 26.49 |
| 70-79 | 668 | 147 | 815 | 81.96 | 18.04 |
| 80-89 | 115 | 16 | 131 | 87.79 | 12.21 |
| 90-99 | 42 | 13 | 55 | 76.36 | 23.64 |
| Total | 37155 | 11687 | 48842 | 76.07 | 23.93 |
When you set return_dict=True, you can access these crosstabs as
DataFrames by assigning them to their own vriables. For example:
crosstab_age_sex = stacked_crosstabs["sex"]
crosstab_age_income = stacked_crosstabs["income"]
Pivoted Stacked Bar Plots Example
Using the census dataset [1], to create horizontal stacked bar plots, set the kind parameter to
"barh" in the stacked_crosstab_plot function. This option pivots the
standard vertical stacked bar plot into a horizontal orientation, making it easier
to compare categories when there are many labels on the y-axis.
Non-Normalized Stacked Bar Plots Example
In the census data [1], to create stacked bar plots without the normalized versions,
set the plot_type parameter to "regular" in the stacked_crosstab_plot
function. This option removes the display of normalized plots beneath the regular
versions. Alternatively, setting the plot_type to "normalized" will display
only the normalized plots. The example below demonstrates regular stacked bar plots
for income by age.
Regular Non-Stacked Bar Plots Example
In the census data [1], to generate regular (non-stacked) bar plots without
displaying their normalized versions, set the plot_type parameter to "regular"
in the stacked_crosstab_plot function and enable remove_stacks by setting
it to True. This configuration removes any stacked elements and prevents the
display of normalized plots beneath the regular versions. Alternatively, setting
plot_type to "normalized" will display only the normalized plots.
When unstacking bar plots in this fashion, the distribution is aligned in descending order, making it easier to visualize the most prevalent categories.
In the example below, the color of the bars has been set to a dark grey (#333333),
and the legend has been removed by setting show_legend=False. This illustrates
regular bar plots for income by age, without stacking.
Box and Violin Plots
Create and save individual boxplots or violin plots, an entire grid of plots, or both for specified metrics and comparisons.
The box_violin_plot function generates individual and/or grid-based plots of boxplots or violin plots for specified metrics against comparison categories in a DataFrame. It offers extensive customization options, including control over plot type, display mode, axis label rotation, figure size, and saving preferences, making it suitable for a wide range of data visualization needs.
This function supports: - Rotating plots (swapping x and y axes). - Adjusting font sizes for axis labels and tick labels. - Wrapping plot titles for better readability. - Saving plots in PNG and/or SVG format with customizable file paths. - Visualizing the distribution of metrics across categories, either individually, as a grid, or both.
- box_violin_plot(df, metrics_list, metrics_comp, n_rows=None, n_cols=None, image_path_png=None, image_path_svg=None, save_plots=False, show_legend=True, plot_type='boxplot', xlabel_rot=0, show_plot='both', rotate_plot=False, individual_figsize=(6, 4), grid_figsize=None, label_fontsize=12, tick_fontsize=10, text_wrap=50, xlim=None, ylim=None, label_names=None, **kwargs)
- Parameters:
df (pandas.DataFrame) – The DataFrame containing the data to plot.
metrics_list (list of str) – List of column names representing the metrics to plot.
metrics_comp (list of str) – List of column names representing the comparison categories.
n_rows (int, optional) – Number of rows in the subplot grid. Automatically calculated if not provided.
n_cols (int, optional) – Number of columns in the subplot grid. Automatically calculated if not provided.
image_path_png (str, optional) – Directory path to save plots in PNG format.
image_path_svg (str, optional) – Directory path to save plots in SVG format.
save_plots (bool, optional) – Boolean indicating whether to save plots. Defaults to
False.show_legend (bool, optional) – Whether to display the legend in the plots. Defaults to
True.plot_type (str, optional) – Type of plot to generate, either
"boxplot"or"violinplot". Defaults to"boxplot".xlabel_rot (int, optional) – Rotation angle for x-axis labels. Defaults to
0.show_plot (str, optional) – Specify the plot display mode:
"individual","grid", or"both". Defaults to"both".rotate_plot (bool, optional) – Whether to rotate the plots by swapping the x and y axes. Defaults to
False.individual_figsize (tuple, optional) – Dimensions (width, height) for individual plots. Defaults to
(6, 4).grid_figsize (tuple, optional) – Dimensions (width, height) for the grid plot.
label_fontsize (int, optional) – Font size for axis labels. Defaults to
12.tick_fontsize (int, optional) – Font size for tick labels. Defaults to
10.text_wrap (int, optional) – Maximum width of plot titles before wrapping. Defaults to
50.xlim (tuple or list, optional) – Limits for the x-axis as a tuple or list (
min,max).ylim (tuple or list, optional) – Limits for the y-axis as a tuple or list (
min,max).label_names (dict, optional) – Dictionary mapping original column names to custom labels for display purposes.
kwargs (additional keyword arguments) – Additional keyword arguments passed to the Seaborn plotting function.
- Raises:
If
show_plotis not one of"individual","grid", or"both".If
save_plotsisTruebut neitherimage_path_pngnorimage_path_svgis specified.If
rotate_plotis not a boolean value.If
individual_figsizeis not a tuple or list of two numbers.If
grid_figsizeis provided and is not a tuple or list of two numbers.
- Returns:
None
Notes
Automatically calculates grid dimensions if
n_rowsandn_colsare not specified.Rotating plots swaps the roles of the x and y axes.
Saving plots requires specifying valid file paths for PNG and/or SVG formats.
Supports customization of plot labels, title wrapping, and font sizes for publication-quality visuals.
This function provides the ability to create and save boxplots or violin plots for specified metrics and comparison categories. It supports the generation of individual plots, a grid of plots, or both. Users can customize the appearance, save the plots to specified directories, and control the display of legends and labels.
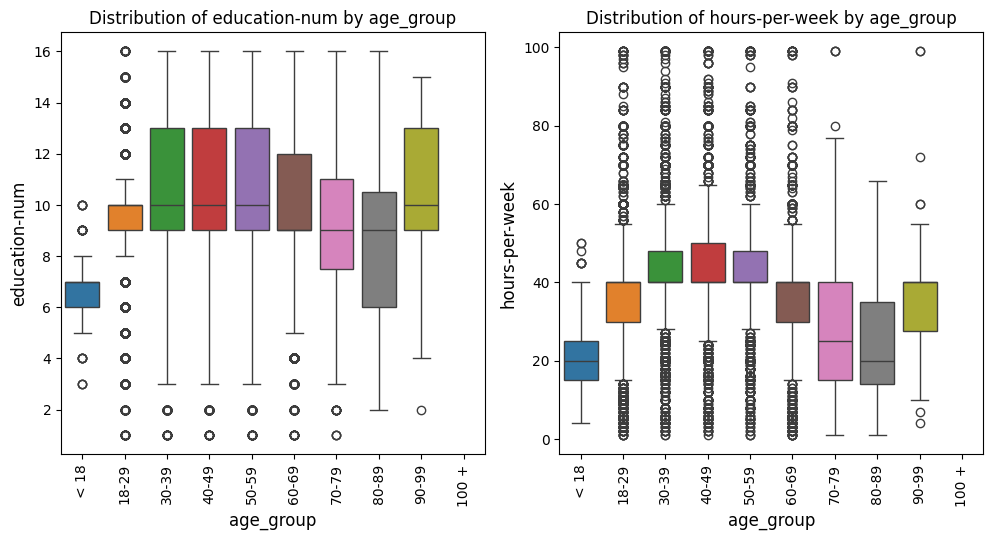
Box Plots Grid Example
In this example with the US census data [1], the box_violin_plot function is employed to create a grid of
boxplots, comparing different metrics against the "age_group" column in the
DataFrame. The metrics_comp parameter is set to ["age_group"], meaning
that the comparison will be based on different age groups. The metrics_list is
provided as age_boxplot_list, which contains the specific metrics to be visualized.
The function is configured to arrange the plots in a grid formatThe image_path_png and
image_path_svg parameters are specified to save the plots in both PNG and
SVG formats, and the save_plots option is set to "all", ensuring that both
individual and grid plots are saved.
The plots are displayed in a grid format, as indicated by the show_plot="grid"
parameter. The plot_type is set to "boxplot", so the function will generate
boxplots for each metric in the list. Additionally, the `x-axis` labels are rotated
by 90 degrees (xlabel_rot=90) to ensure that the labels are legible. The legend is
hidden by setting show_legend=False, keeping the plots clean and focused on the data.
This configuration provides a comprehensive visual comparison of the specified
metrics across different age groups, with all plots saved for future reference or publication.
age_boxplot_list = df[
[
"education-num",
"hours-per-week",
]
].columns.to_list()
from eda_toolkit import box_violin_plot
metrics_comp = ["age_group"]
box_violin_plot(
df=df,
metrics_list=age_boxplot_list,
metrics_comp=metrics_comp,
image_path_png=image_path_png,
image_path_svg=image_path_svg,
save_plots="all",
show_plot="both",
show_legend=False,
plot_type="boxplot",
xlabel_rot=90,
)
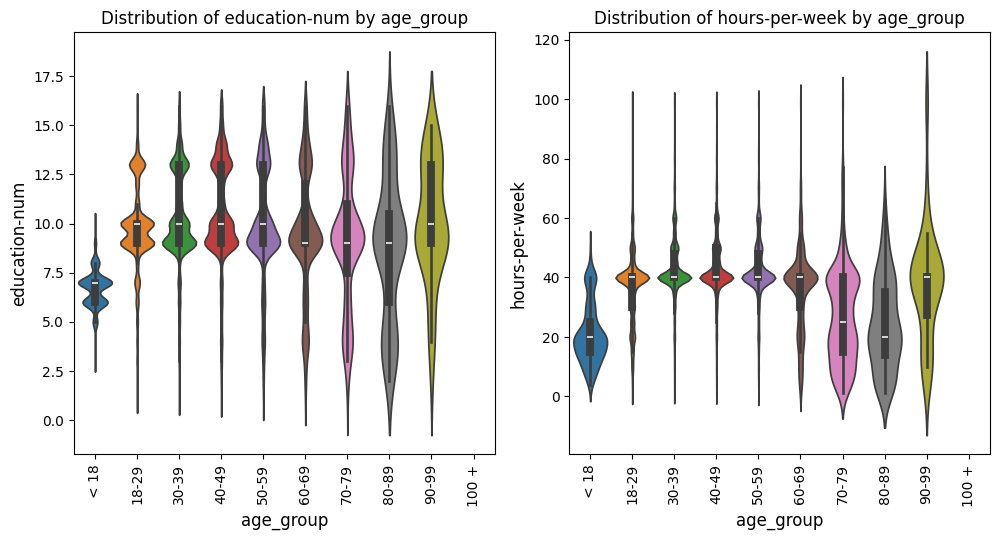
Violin Plots Grid Example
In this example with the US census data [1], we keep everything the same as the prior example, but change the
plot_type to violinplot. This adjustment will generate violin plots instead
of boxplots while maintaining all other settings.
from eda_toolkit import box_violin_plot
metrics_comp = ["age_group"]
box_violin_plot(
df=df,
metrics_list=age_boxplot_list,
metrics_comp=metrics_comp,
image_path_png=image_path_png,
image_path_svg=image_path_svg,
save_plots="all",
show_plot="both",
show_legend=False,
plot_type="violinplot",
xlabel_rot=90,
)
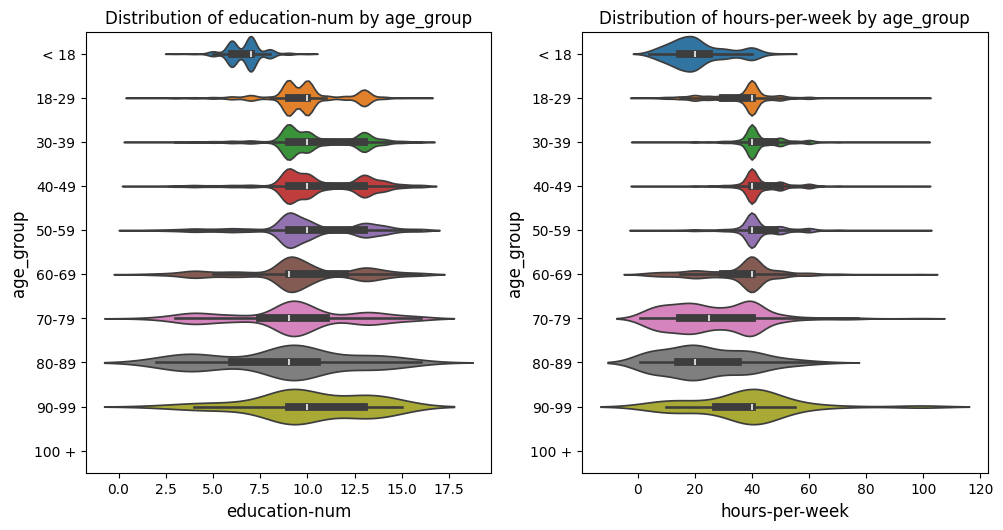
Pivoted Violin Plots Grid Example
In this example with the US census data [1], we set xlabel_rot=0 and rotate_plot=True
to pivot the plot, changing the orientation of the axes while keeping the x-axis labels upright.
This adjustment flips the axes, providing a different perspective on the data distribution.
from eda_toolkit import box_violin_plot
metrics_comp = ["age_group"]
box_violin_plot(
df=df,
metrics_list=age_boxplot_list,
metrics_comp=metrics_comp,
show_plot="both",
rotate_plot=True,
show_legend=False,
plot_type="violinplot",
xlabel_rot=0,
)
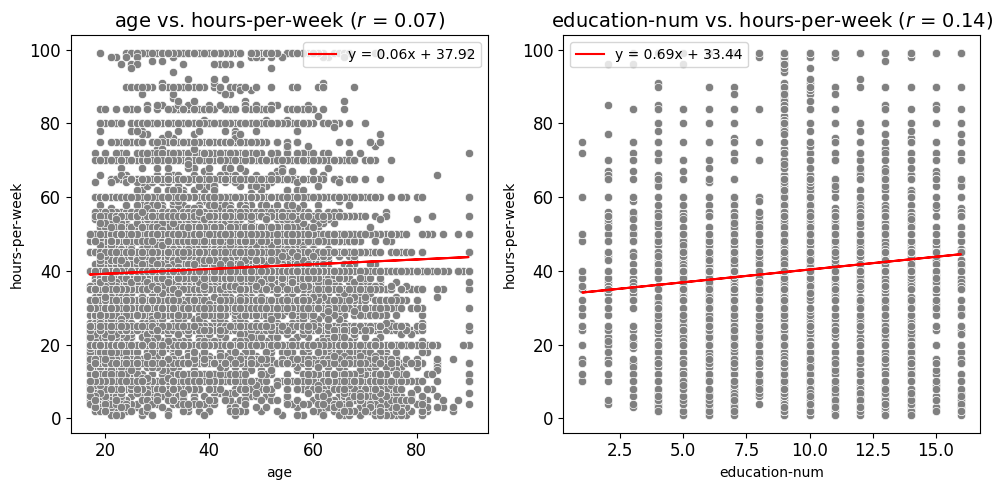
Scatter Plots and Best Fit Lines
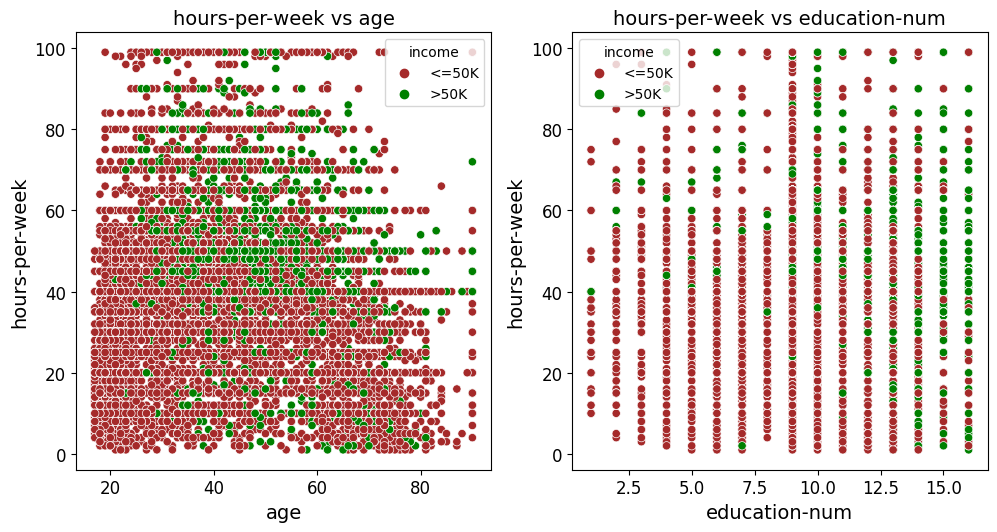
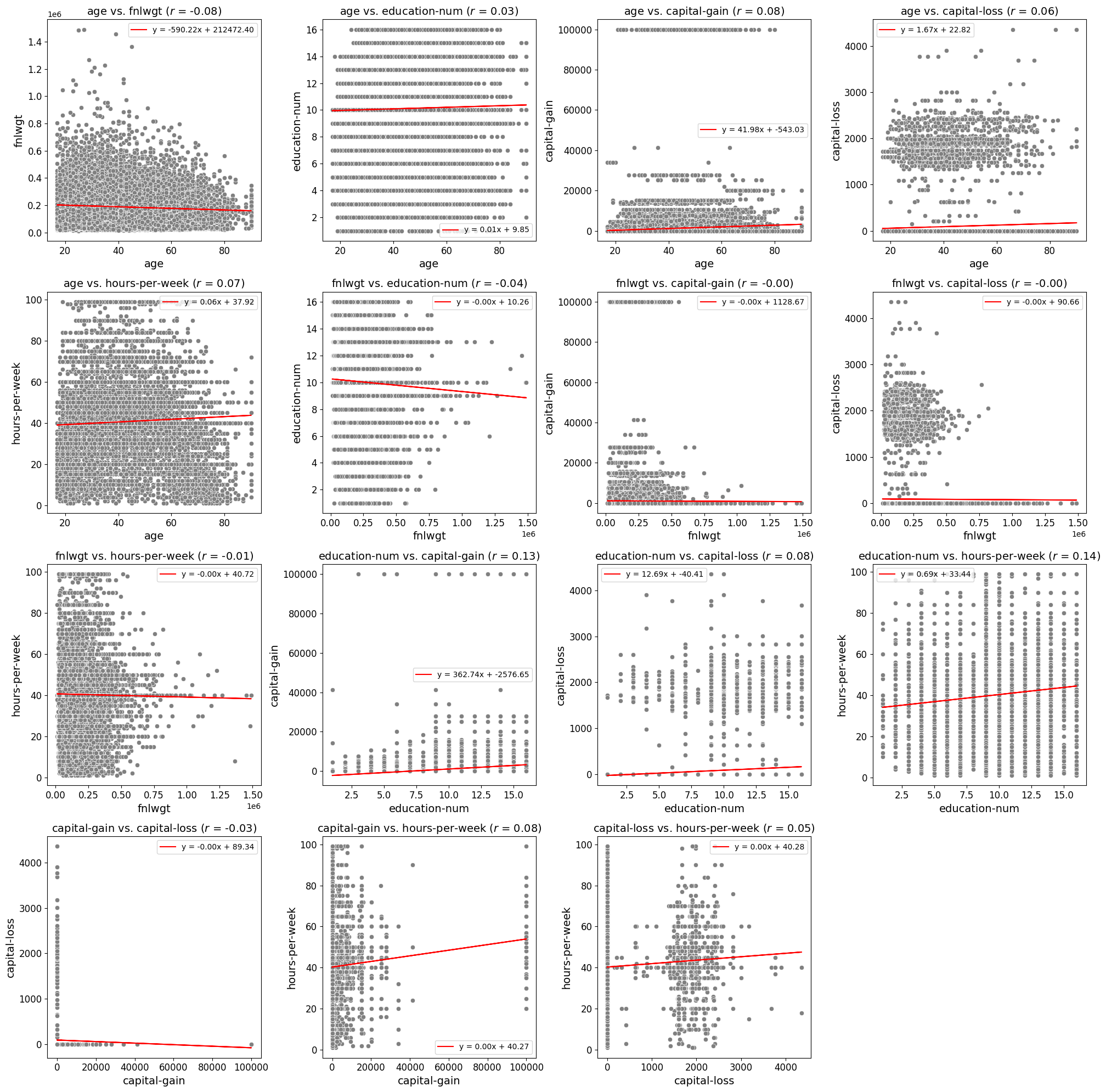
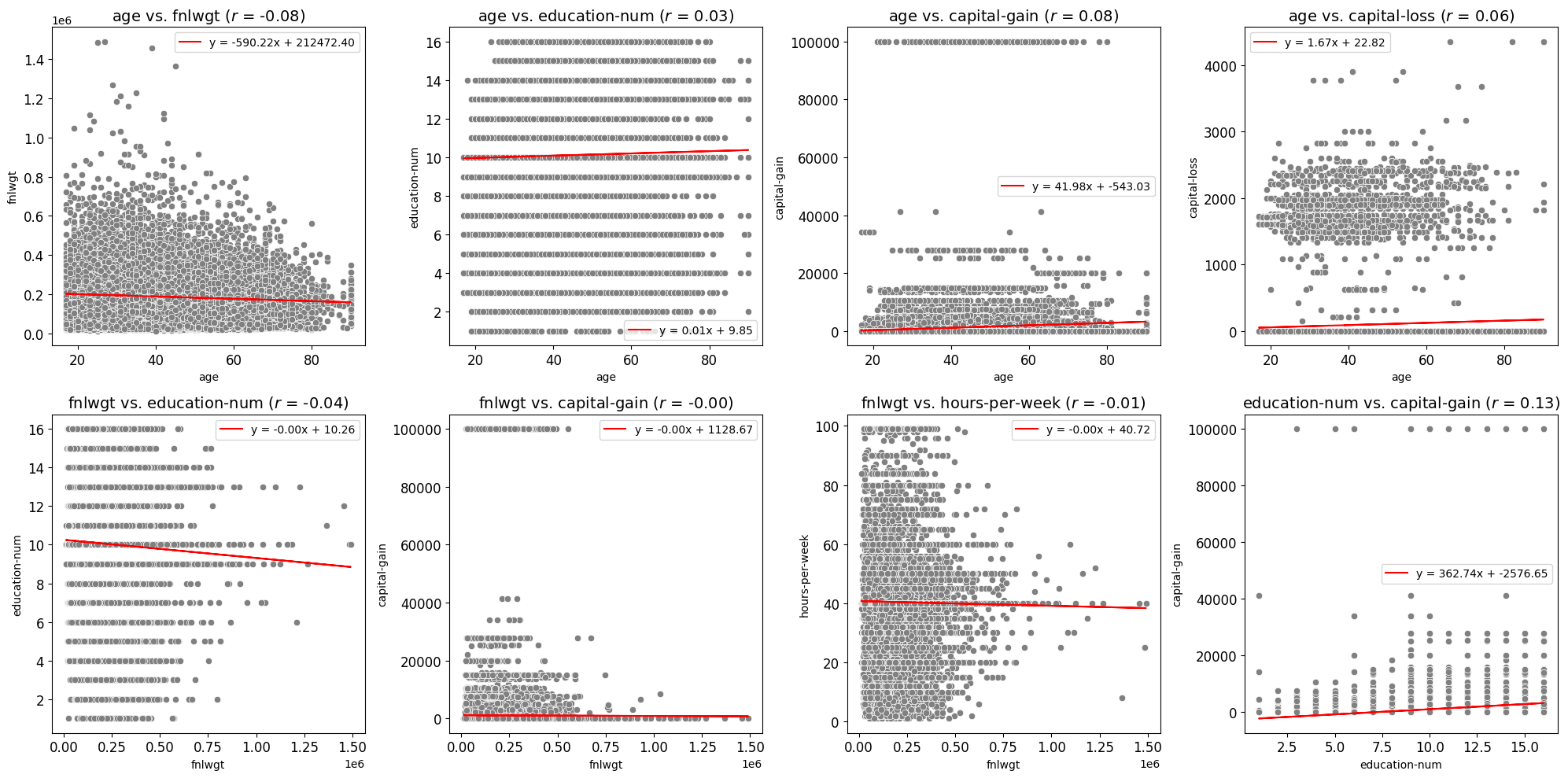
Scatter Fit Plot
Create and Save Scatter Plots or a Grid of Scatter Plots
This function, scatter_fit_plot, generates scatter plots for one or more pairs of variables (x_vars and y_vars) from a given DataFrame. The function can produce individual scatter plots or organize multiple scatter plots into a grid layout, enabling easy visualization of relationships between different pairs of variables in a unified view.
Optional Features
Best Fit Line: Adds an optional best fit line to the scatter plots, calculated using linear regression. This provides visual insights into the linear relationship between variables. The equation of the line can also be displayed in the plot’s legend.
Correlation Coefficient Display: Displays the Pearson correlation coefficient directly in the plot title, offering a numeric indicator of the linear relationship between variables.
Customization Options
The function provides a variety of parameters to customize the appearance and behavior of scatter plots:
Color: Customize point colors with a fixed color or a
hueparameter to group points by a categorical variable.Size and Marker: Adjust the size of points dynamically based on a variable, and specify custom markers to represent data effectively.
Labels and Axis Configuration: Set axis labels, font sizes, label rotation, and axis limits to ensure readability and focus on relevant data.
Plot Management
Display Options: Control how plots are displayed, whether as individual plots, in a grid, or both.
Saving Options: Save the generated plots in PNG or SVG formats. Specify which plots to save (e.g.,
"all","individual", or"grid") and their respective file paths.
Advanced Features
Combination Plotting: Automatically generate scatter plots for all combinations of variables when using the
all_varsparameter.Exclusion Rules: Exclude specific (
x_var,y_var) combinations from being plotted using theexclude_combinationsparameter.Legend and Aesthetic Adjustments: Toggle legends, adjust plot size for individual or grid plots, and wrap long titles for clarity.
Error Handling
The function performs comprehensive input validation to prevent common errors, such as:
Conflicting inputs for
all_vars,x_vars, andy_vars.Invalid or missing column names in the DataFrame.
Incorrectly specified parameters like axis limits or figure sizes.
- scatter_fit_plot(df, x_vars=None, y_vars=None, all_vars=None, exclude_combinations=None, n_rows=None, n_cols=None, max_cols=4, image_path_png=None, image_path_svg=None, save_plots=None, show_legend=True, xlabel_rot=0, show_plot='grid', rotate_plot=False, individual_figsize=(6, 4), grid_figsize=None, label_fontsize=12, tick_fontsize=10, text_wrap=50, add_best_fit_line=False, scatter_color='C0', best_fit_linecolor='red', best_fit_linestyle='-', hue=None, hue_palette=None, size=None, sizes=None, marker='o', show_correlation=True, xlim=None, ylim=None, label_names=None, **kwargs)
Generate scatter plots or a grid of scatter plots for the given
x_varsandy_vars, with optional best fit lines, correlation coefficients, and customizable aesthetics.- Parameters:
df (pandas.DataFrame) – The DataFrame containing the data for the plots.
x_vars (list of str or str, optional) – List of variable names to plot on the
x-axis. If a single string is provided, it will be converted into a list with one element.y_vars (list of str or str, optional) – List of variable names to plot on the
y-axis. If a single string is provided, it will be converted into a list with one element.all_vars (list of str, optional) – Generates scatter plots for all combinations of variables in this list, overriding
x_varsandy_vars.exclude_combinations (list of tuples, optional) – List of (
x_var,y_var) combinations to exclude from plotting.n_rows (int, optional) – Number of rows in the subplot grid. Calculated automatically if not specified.
n_cols (int, optional) – Number of columns in the subplot grid. Calculated automatically if not specified.
max_cols (int, optional) – Maximum number of columns in the subplot grid. Default is
4.image_path_png (str, optional) – Directory path to save PNG images of scatter plots.
image_path_svg (str, optional) – Directory path to save SVG images of scatter plots.
save_plots (str, optional) – Specify which plots to save (
"all","individual", or"grid"); uses a progress bar (tqdm) for saving.show_legend (bool, optional) – Toggle display of the plot legend. Default is
True.xlabel_rot (int, optional) – Angle to rotate
x-axislabels. Default is0.show_plot (str, optional) – Controls plot display:
"individual","grid","both", or"combinations". Default is"grid". Use"combinations"to return all valid (x, y) variable pairs without generating plots.rotate_plot (bool, optional) – Rotate plots (swap
xandyaxes). Default isFalse.individual_figsize (tuple or list, optional) – Dimensions for individual plot figures (width, height). Default is
(6, 4).grid_figsize (tuple or list, optional) – Dimensions for grid plots (width, height). Calculated automatically if not specified.
label_fontsize (int, optional) – Font size for axis labels. Default is
12.tick_fontsize (int, optional) – Font size for axis ticks. Default is
10.text_wrap (int, optional) – Maximum title width before wrapping. Default is
50.add_best_fit_line (bool, optional) – Add a best fit line to scatter plots. Default is
False.scatter_color (str, optional) – Default color for scatter points. Default is
"C0".best_fit_linecolor (str, optional) – Color for the best fit line. Default is
"red".best_fit_linestyle (str, optional) – Linestyle for the best fit line. Default is
"-".hue (str, optional) – Column name for grouping variable to color points differently.
hue_palette (dict, list, or str, optional) – Colors for each
huelevel (requireshue).size (str, optional) – Variable to scale scatter point sizes.
sizes (dict, optional) – Mapping of minimum and maximum point sizes.
marker (str, optional) – Style of scatter points. Default is
"o".show_correlation (bool, optional) – Display correlation coefficient in titles. Default is
True.xlim (tuple or list, optional) – Limits for the
x-axis(min,max).ylim (tuple or list, optional) – Limits for the
y-axis(min,max).label_names (dict, optional) – Rename columns for display in titles and labels.
kwargs (dict, optional) – Additional options for
sns.scatterplot.
- Raises:
ValueError – See function docstring for detailed error conditions.
- Returns:
None. Generates and optionally saves scatter plots.
Regression-Centric Scatter Plots Example
In this US census data [1] example, the scatter_fit_plot function is
configured to display the Pearson correlation coefficient and a best fit line
on each scatter plot. The correlation coefficient is shown in the plot title,
controlled by the show_correlation=True parameter, which provides a measure
of the strength and direction of the linear relationship between the variables.
Additionally, the add_best_fit_line=True parameter adds a best fit line to
each plot, with the equation for the line displayed in the legend. This equation,
along with the best fit line, helps to visually assess the relationship between
the variables, making it easier to identify trends and patterns in the data. The
combination of the correlation coefficient and the best fit line offers both
a quantitative and visual representation of the relationships, enhancing the
interpretability of the scatter plots.
from eda_toolkit import scatter_fit_plot
scatter_fit_plot(
df=df,
x_vars=["age", "education-num"],
y_vars=["hours-per-week"],
show_legend=True,
show_plot="grid",
grid_figsize=None,
label_fontsize=14,
tick_fontsize=12,
add_best_fit_line=True,
scatter_color="#808080",
show_correlation=True,
)
Scatter Plots Grouped by Category Example
In this example, the scatter_fit_plot function is used to generate a grid of
scatter plots that examine the relationships between age and hours-per-week
as well as education-num and hours-per-week. Compared to the previous
example, a few key inputs have been changed to adjust the appearance and functionality
of the plots:
Hue and Hue Palette: The
hueparameter is set to"income", meaning that the data points in the scatter plots are colored according to the values in theincomecolumn. A custom color mapping is provided via thehue_paletteparameter, where the income categories"<=50K"and">50K"are assigned the colors"brown"and"green", respectively. This change visually distinguishes the data points based on income levels.Scatter Color: The
scatter_colorparameter is set to"#808080", which applies a grey color to the scatter points when nohueis provided. However, since ahueis specified in this example, thehue_palettetakes precedence and overrides this color setting.Best Fit Line: The
add_best_fit_lineparameter is set toFalse, meaning that no best fit line is added to the scatter plots. This differs from the previous example where a best fit line was included.Correlation Coefficient: The
show_correlationparameter is set toFalse, so the Pearson correlation coefficient will not be displayed in the plot titles. This is another change from the previous example where the correlation coefficient was included.Hue Legend: The
show_legendparameter remains set toTrue, ensuring that the legend displaying the hue categories ("<=50K"and">50K") appears on the plots, helping to interpret the color coding of the data points.
These changes allow for the creation of scatter plots that highlight the income levels of individuals, with custom color coding and without additional elements like a best fit line or correlation coefficient. The resulting grid of plots is then saved as images in the specified paths.
from eda_toolkit import scatter_fit_plot
hue_dict = {"<=50K": "brown", ">50K": "green"}
scatter_fit_plot(
df=df,
x_vars=["age", "education-num"],
y_vars=["hours-per-week"],
show_legend=True,
show_plot="grid",
label_fontsize=14,
tick_fontsize=12,
add_best_fit_line=False,
scatter_color="#808080",
hue="income",
hue_palette=hue_dict,
show_correlation=False,
)
Scatter Plots (All Combinations Example)
In this example, the scatter_fit_plot function is used to generate a grid of scatter plots that explore the relationships between all numeric variables in the df DataFrame. The function automatically identifies and plots all possible combinations of these variables. Below are key aspects of this example:
All Variables Combination: The
all_varsparameter is used to automatically generate scatter plots for all possible combinations of numerical variables in the DataFrame. This means you don’t need to manually specifyx_varsandy_vars, as the function will iterate through each possible pair.Grid Display: The
show_plotparameter is set to"grid", so the scatter plots are displayed in a grid format. This is useful for comparing multiple relationships simultaneously.Font Sizes: The
label_fontsizeandtick_fontsizeparameters are set to14and12, respectively. This increases the readability of axis labels and tick marks, making the plots more visually accessible.Best Fit Line: The
add_best_fit_lineparameter is set toTrue, meaning that a best fit line is added to each scatter plot. This helps in visualizing the linear relationship between variables.Scatter Color: The
scatter_colorparameter is set to"#808080", applying a grey color to the scatter points. This provides a neutral color that does not distract from the data itself.Correlation Coefficient: The
show_correlationparameter is set toTrue, so the Pearson correlation coefficient will be displayed in the plot titles. This helps to quantify the strength of the relationship between the variables.
These settings allow for the creation of scatter plots that comprehensively explore the relationships between all numeric variables in the DataFrame. The plots are saved in a grid format, with added best fit lines and correlation coefficients for deeper analysis. The resulting images can be stored in the specified directory for future reference.
from eda_toolkit import scatter_fit_plot
scatter_fit_plot(
df=df,
all_vars=df.select_dtypes(np.number).columns.to_list(),
show_legend=True,
show_plot="grid",
label_fontsize=14,
tick_fontsize=12,
add_best_fit_line=True,
scatter_color="#808080",
show_correlation=True,
)
Scatter Plots: Excluding Specific Combinations
Example 1
In this example, the scatter_fit_plot function is used to generate a grid of
scatter plots while excluding specific combinations of variables. This allows for
a more targeted exploration of relationships, omitting plots that may be redundant
or irrelevant. Below are key aspects of this example:
1. Exclude Combinations: The exclude_combinations parameter is used to
specify pairs of variables that should be excluded from the scatter plot grid.
For example:
("capital-gain", "hours-per-week")
("capital-loss", "capital-gain")
("education-num", "hours-per-week")
This ensures that the generated plots are more relevant to the analysis.
2. All Variables Combination: Like the previous example, the all_vars parameter
is used to automatically generate scatter plots for all combinations of numerical
variables in the DataFrame, minus the excluded pairs.
3. Grid Display: The show_plot parameter is set to "grid", allowing for
simultaneous visualization of multiple relationships in a cohesive layout.
By excluding specific variable combinations, this example demonstrates how to fine-tune the generated scatter plots to focus only on the most meaningful comparisons.
from eda_toolkit import scatter_fit_plot
exclude_combinations = [
("capital-gain", "hours-per-week"),
("capital-loss", "capital-gain"),
("capital-loss", "hours-per-week"),
("capital-loss", "education-num"),
("capital-loss", "fnlwgt"),
("education-num", "hours-per-week"),
("hours-per-week", "age"),
]
scatter_fit_plot(
df=df,
all_vars=df.select_dtypes(np.number).columns.to_list(),
show_legend=True,
exclude_combinations=exclude_combinations,
show_plot="grid",
label_fontsize=14,
tick_fontsize=12,
add_best_fit_line=True,
scatter_color="#808080",
show_correlation=True,
)
Example 2
In this example, the scatter_fit_plot function is used to generate a list of
valid (x, y) combinations without creating any plots. This feature is particularly
useful when identifying potential variable pairs for further analysis or programmatically
excluding irrelevant combinations. Below are key aspects of this example:
1. List of Combinations: Setting the show_plot parameter to "combinations"
returns a list of all valid combinations of numerical variables. The exclude_combinations
parameter is used to omit specific pairs from this list.
Excluded Combinations: Certain variable pairs are excluded using the
exclude_combinationsparameter. For instance:("capital-gain", "hours-per-week")("capital-loss", "education-num")("hours-per-week", "age")
This ensures the returned list only includes meaningful and relevant combinations.
3. Order Irrelevance: The order of variables in each tuple is irrelevant. For example,
("capital-gain", "hours-per-week") and ("hours-per-week", "capital-gain") are
considered the same pair, and only one is included in the output.
4. Efficient Output: No scatter plots are generated or displayed, making this approach efficient for exploratory analysis or preprocessing.
Below is an example illustrating how to retrieve a list of combinations:
from eda_toolkit import scatter_fit_plot
exclude_combinations = [
("capital-gain", "hours-per-week"),
("capital-loss", "capital-gain"),
("capital-loss", "hours-per-week"),
("capital-loss", "education-num"),
("capital-loss", "fnlwgt"),
("education-num", "hours-per-week"),
("hours-per-week", "age"),
]
combinations = scatter_fit_plot(
df=df,
all_vars=df.select_dtypes(np.number).columns.to_list(),
show_legend=True,
exclude_combinations=exclude_combinations,
show_plot="combinations",
label_fontsize=14,
tick_fontsize=12,
add_best_fit_line=True,
scatter_color="#808080",
show_correlation=True,
image_path_png=image_path_png,
image_path_svg=image_path_svg,
save_plots="grid",
)
print(combinations)
The returned list of combinations will look like this:
[
('age', 'fnlwgt'),
('age', 'education-num'),
('age', 'capital-gain'),
('age', 'capital-loss'),
('fnlwgt', 'education-num'),
('fnlwgt', 'capital-gain'),
('fnlwgt', 'hours-per-week'),
('education-num', 'capital-gain'),
]
This feature allows users to explore potential variable relationships before deciding on visualization or further analysis.
Correlation Matrices
Generate and Save Customizable Correlation Heatmaps
The flex_corr_matrix function is designed to create highly customizable correlation heatmaps for visualizing the relationships between variables in a DataFrame. This function allows users to generate either a full or triangular correlation matrix, with options for annotation, color mapping, and saving the plot in multiple formats.
Customizable Plot Appearance
The function provides extensive customization options for the heatmap’s appearance:
Colormap Selection: Choose from a variety of colormaps to represent the strength of correlations. The default is
"coolwarm", but this can be adjusted to fit the needs of the analysis.Annotation: Optionally annotate the heatmap with correlation coefficients, making it easier to interpret the strength of relationships at a glance.
Figure Size and Layout: Customize the dimensions of the heatmap to ensure it fits well within reports, presentations, or dashboards.
Triangular vs. Full Correlation Matrix
A key feature of the flex_corr_matrix function is the ability to generate either a full correlation matrix or only the upper triangle. This option is particularly useful when the matrix is large, as it reduces visual clutter and focuses attention on the unique correlations.
Label and Axis Configuration
The function offers flexibility in configuring axis labels and titles:
Label Rotation: Rotate x-axis and y-axis labels for better readability, especially when working with long variable names.
Font Sizes: Adjust the font sizes of labels and tick marks to ensure the plot is clear and readable.
Title Wrapping: Control the wrapping of long titles to fit within the plot without overlapping other elements.
Plot Display and Saving Options
The flex_corr_matrix function allows you to display the heatmap directly or save it as PNG or SVG files for use in reports or presentations. If saving is enabled, you can specify file paths and names for the images.
- flex_corr_matrix(df, cols=None, annot=True, cmap='coolwarm', save_plots=False, image_path_png=None, image_path_svg=None, figsize=(10, 10), title=None, label_fontsize=12, tick_fontsize=10, xlabel_rot=45, ylabel_rot=0, xlabel_alignment='right', ylabel_alignment='center_baseline', text_wrap=50, vmin=-1, vmax=1, cbar_label='Correlation Index', triangular=True, **kwargs)
Create a customizable correlation heatmap with options for annotation, color mapping, figure size, and saving the plot.
- Parameters:
df (pandas.DataFrame) – The DataFrame containing the data.
cols (list of str, optional) – List of column names to include in the correlation matrix. If None, all columns are included.
annot (bool, optional) – Whether to annotate the heatmap with correlation coefficients. Default is
True.cmap (str, optional) – The colormap to use for the heatmap. Default is
"coolwarm".save_plots (bool, optional) – Controls whether to save the plots. Default is
False.image_path_png (str, optional) – Directory path to save PNG images of the heatmap.
image_path_svg (str, optional) – Directory path to save SVG images of the heatmap.
figsize (tuple, optional) – Width and height of the figure for the heatmap. Default is
(10, 10).title (str, optional) – Title of the heatmap. Default is
None.label_fontsize (int, optional) – Font size for tick labels and colorbar label. Default is
12.tick_fontsize (int, optional) – Font size for axis tick labels. Default is
10.xlabel_rot (int, optional) – Rotation angle for x-axis labels. Default is
45.ylabel_rot (int, optional) – Rotation angle for y-axis labels. Default is
0.xlabel_alignment (str, optional) – Horizontal alignment for x-axis labels. Default is
"right".ylabel_alignment (str, optional) – Vertical alignment for y-axis labels. Default is
"center_baseline".text_wrap (int, optional) – The maximum width of the title text before wrapping. Default is
50.vmin (float, optional) – Minimum value for the heatmap color scale. Default is
-1.vmax (float, optional) – Maximum value for the heatmap color scale. Default is
1.cbar_label (str, optional) – Label for the colorbar. Default is
"Correlation Index".triangular (bool, optional) – Whether to show only the upper triangle of the correlation matrix. Default is
True. Excludes the diagonal and lower triangle if enabled.kwargs (dict, optional) – Additional keyword arguments for
seaborn.heatmap()to customize the plot further.
- Raises:
If
annotis not a boolean.If
colsis not a list of column names.If
save_plotsis not a boolean.If
triangularis not a boolean.If
save_plotsis True but neitherimage_path_pngnorimage_path_svgis specified.
- Returns:
NoneThis function does not return any value but generates and optionally saves a correlation heatmap.
Note
To save images, you must specify the paths for image_path_png or image_path_svg.
Saving plots is triggered by providing a valid save_formats string.
Triangular Correlation Matrix Example
The provided code filters the census [1] DataFrame df to include only numeric columns using
select_dtypes(np.number). It then utilizes the flex_corr_matrix() function
to generate a right triangular correlation matrix, which only displays the
upper half of the correlation matrix. The heatmap is customized with specific
colormap settings, title, label sizes, axis label rotations, and other formatting
options.
Note
This triangular matrix format is particularly useful for avoiding redundancy in correlation matrices, as it excludes the lower half, making it easier to focus on unique pairwise correlations.
The function also includes a labeled color bar, helping users quickly interpret the strength and direction of the correlations.
# Select only numeric data to pass into the function
df_num = df.select_dtypes(np.number)
from eda_toolkit import flex_corr_matrix
flex_corr_matrix(
df=df,
cols=df_num.columns.to_list(),
annot=True,
cmap="coolwarm",
figsize=(10, 8),
title="US Census Correlation Matrix",
xlabel_alignment="right",
label_fontsize=14,
tick_fontsize=12,
xlabel_rot=45,
ylabel_rot=0,
text_wrap=50,
vmin=-1,
vmax=1,
cbar_label="Correlation Index",
triangular=True,
)
Full Correlation Matrix Example
In this modified census [1] example, the key changes are the use of the viridis
colormap and the decision to plot the full correlation matrix instead of just the
upper triangle. By setting cmap="viridis", the heatmap will use a different
color scheme, which can provide better visual contrast or align with specific
aesthetic preferences. Additionally, by setting triangular=False, the full
correlation matrix is displayed, allowing users to view all pairwise correlations,
including both upper and lower halves of the matrix. This approach is beneficial
when you want a comprehensive view of all correlations in the dataset.
from eda_toolkit import flex_corr_matrix
flex_corr_matrix(
df=df,
cols=df_num.columns.to_list(),
annot=True,
cmap="viridis",
figsize=(10, 8),
title="US Census Correlation Matrix",
xlabel_alignment="right",
label_fontsize=14,
tick_fontsize=12,
xlabel_rot=45,
ylabel_rot=0,
text_wrap=50,
vmin=-1,
vmax=1,
cbar_label="Correlation Index",
triangular=False,
)
Partial Dependence Plots
Partial Dependence Plots (PDPs) are a powerful tool in machine learning interpretability, providing insights into how features influence the predicted outcome of a model. PDPs can be generated in both 2D and 3D, depending on whether you want to analyze the effect of one feature or the interaction between two features on the model’s predictions.
2D Partial Dependence Plots
The plot_2d_pdp function generates 2D partial dependence plots (PDPs) for specified features or pairs of features. These plots help analyze the marginal effect of individual or paired features on the predicted outcome.
Key Features:
Flexible Plot Layouts: Generate all 2D PDPs in a grid layout, as separate individual plots, or both for maximum versatility.
Customization Options: Adjust figure size, font sizes for labels and ticks, and wrap long titles to ensure clear and visually appealing plots.
Save Plots: Save generated plots in PNG or SVG formats with options to save all plots, only individual plots, or just the grid plot.
- plot_2d_pdp(model, X_train, feature_names, features, title='Partial dependence plot', grid_resolution=50, plot_type='grid', grid_figsize=(12, 8), individual_figsize=(6, 4), label_fontsize=12, tick_fontsize=10, text_wrap=50, image_path_png=None, image_path_svg=None, save_plots=None, file_prefix='partial_dependence')
Generate and save 2D partial dependence plots for specified features using a trained machine learning model. The function supports grid and individual layouts and provides options for customization and saving plots in various formats.
- Parameters:
model (estimator object) – The trained machine learning model used to generate partial dependence plots.
X_train (pandas.DataFrame or numpy.ndarray) – The training data used to compute partial dependence. Should correspond to the features used to train the model.
feature_names (list of str) – A list of feature names corresponding to the columns in
X_train.features (list of int or tuple of int) – A list of feature indices or tuples of feature indices for which to generate partial dependence plots.
title (str, optional) – The title for the entire plot. Default is
"Partial dependence plot".grid_resolution (int, optional) – The resolution of the grid used to compute the partial dependence. Higher values provide smoother curves but may increase computation time. Default is
50.plot_type (str, optional) – The type of plot to generate. Choose
"grid"for a grid layout,"individual"for separate plots, or"both"to generate both layouts. Default is"grid".grid_figsize (tuple, optional) – Tuple specifying the width and height of the figure for the grid layout. Default is
(12, 8).individual_figsize (tuple, optional) – Tuple specifying the width and height of the figure for individual plots. Default is
(6, 4).label_fontsize (int, optional) – Font size for the axis labels and titles. Default is
12.tick_fontsize (int, optional) – Font size for the axis tick labels. Default is
10.text_wrap (int, optional) – The maximum width of the title text before wrapping. Useful for managing long titles. Default is
50.image_path_png (str, optional) – The directory path where PNG images of the plots will be saved, if saving is enabled.
image_path_svg (str, optional) – The directory path where SVG images of the plots will be saved, if saving is enabled.
save_plots (str, optional) – Controls whether to save the plots. Options include
"all","individual","grid", orNone(default). If saving is enabled, ensureimage_path_pngorimage_path_svgare provided.file_prefix (str, optional) – Prefix for the filenames of the saved grid plots. Default is
"partial_dependence".
- Raises:
If
plot_typeis not one of"grid","individual", or"both".If
save_plotsis enabled but neitherimage_path_pngnorimage_path_svgis provided.
- Returns:
NoneThis function generates partial dependence plots and displays them. It does not return any values.
2D Plots - CA Housing Example
Consider a scenario where you have a machine learning model predicting median
house values in California. [4] Suppose you want to understand how non-location
features like the average number of occupants per household (AveOccup) and the
age of the house (HouseAge) jointly influence house values. A 2D partial
dependence plot allows you to visualize this relationship in two ways: either as
individual plots for each feature or as a combined plot showing the interaction
between two features.
For instance, the 2D partial dependence plot can help you analyze how the age of the house impacts house values while holding the number of occupants constant, or vice versa. This is particularly useful for identifying the most influential features and understanding how changes in these features might affect the predicted house value.
If you extend this to two interacting features, such as AveOccup and HouseAge,
you can explore their combined effect on house prices. The plot can reveal how
different combinations of occupancy levels and house age influence the value,
potentially uncovering non-linear relationships or interactions that might not be
immediately obvious from a simple 1D analysis.
Here’s how you can generate and visualize these 2D partial dependence plots using the California housing dataset:
Fetch The CA Housing Dataset and Prepare The DataFrame
from sklearn.datasets import fetch_california_housing
from sklearn.model_selection import train_test_split
from sklearn.ensemble import GradientBoostingRegressor
import pandas as pd
# Load the dataset
data = fetch_california_housing()
df = pd.DataFrame(data.data, columns=data.feature_names)
Split The Data Into Training and Testing Sets
X_train, X_test, y_train, y_test = train_test_split(
df, data.target, test_size=0.2, random_state=42
)
Train a GradientBoostingRegressor Model
model = GradientBoostingRegressor(
n_estimators=100,
max_depth=4,
learning_rate=0.1,
loss="huber",
random_state=42,
)
model.fit(X_train, y_train)
Create 2D Partial Dependence Plot Grid
from eda_toolkit import plot_2d_pdp
# Feature names
names = data.feature_names
# Generate 2D partial dependence plots
plot_2d_pdp(
model=model,
X_train=X_train,
feature_names=names,
features=[
"MedInc",
"AveOccup",
"HouseAge",
"AveRooms",
"Population",
("AveOccup", "HouseAge"),
],
title="PDP of house value on CA non-location features",
grid_figsize=(14, 10),
individual_figsize=(12, 4),
label_fontsize=14,
tick_fontsize=12,
text_wrap=120,
plot_type="grid",
image_path_png="path/to/save/png",
save_plots="all",
)
3D Partial Dependence Plots
The plot_3d_pdp function extends the concept of partial dependence to three dimensions, allowing you to visualize the interaction between two features and their combined effect on the model’s predictions.
Interactive and Static 3D Plots: Generate static 3D plots using Matplotlib or interactive 3D plots using Plotly. The function also allows for generating both types simultaneously.
Colormap and Layout Customization: Customize the colormaps for both Matplotlib and Plotly plots. Adjust figure size, camera angles, and zoom levels to create plots that fit perfectly within your presentation or report.
Axis and Title Configuration: Customize axis labels for both Matplotlib and Plotly plots. Adjust font sizes and control the wrapping of long titles to maintain readability.
- plot_3d_pdp(model, dataframe, feature_names_list, x_label=None, y_label=None, z_label=None, title, html_file_path=None, html_file_name=None, image_filename=None, plot_type="both", matplotlib_colormap=None, plotly_colormap="Viridis", zoom_out_factor=None, wireframe_color=None, view_angle=(22, 70), figsize=(7, 4.5), text_wrap=50, horizontal=-1.25, depth=1.25, vertical=1.25, cbar_x=1.05, cbar_thickness=25, title_x=0.5, title_y=0.95, top_margin=100, image_path_png=None, image_path_svg=None, show_cbar=True, grid_resolution=20, left_margin=20, right_margin=65, label_fontsize=8, tick_fontsize=6, enable_zoom=True, show_modebar=True)
Generate 3D partial dependence plots for two features of a machine learning model.
This function supports both static (Matplotlib) and interactive (Plotly) visualizations, allowing for flexible and comprehensive analysis of the relationship between two features and the target variable in a model.
- Parameters:
model (estimator object) – The trained machine learning model used to generate partial dependence plots.
dataframe (pandas.DataFrame or numpy.ndarray) – The dataset on which the model was trained or a representative sample. If a DataFrame is provided,
feature_names_listshould correspond to the column names. If a NumPy array is provided,feature_names_listshould correspond to the indices of the columns.feature_names_list (list of str) – A list of two feature names or indices corresponding to the features for which partial dependence plots are generated.
x_label (str, optional) – Label for the x-axis in the plots. Default is
None.y_label (str, optional) – Label for the y-axis in the plots. Default is
None.z_label (str, optional) – Label for the z-axis in the plots. Default is
None.title (str) – The title for the plots.
html_file_path (str, optional) – Path to save the interactive Plotly HTML file. Required if
plot_typeis"interactive"or"both". Default isNone.html_file_name (str, optional) – Name of the HTML file to save the interactive Plotly plot. Required if
plot_typeis"interactive"or"both". Default isNone.image_filename (str, optional) – Base filename for saving static Matplotlib plots as PNG and/or SVG. Default is
None.plot_type (str, optional) – The type of plots to generate. Options are: -
"static": Generate only static Matplotlib plots. -"interactive": Generate only interactive Plotly plots. -"both": Generate both static and interactive plots. Default is"both".matplotlib_colormap (matplotlib.colors.Colormap, optional) – Custom colormap for the Matplotlib plot. If not provided, a default colormap is used.
plotly_colormap (str, optional) – Colormap for the Plotly plot. Default is
"Viridis".zoom_out_factor (float, optional) – Factor to adjust the zoom level of the Plotly plot. Default is
None.wireframe_color (str, optional) – Color for the wireframe in the Matplotlib plot. If
None, no wireframe is plotted. Default isNone.view_angle (tuple, optional) – Elevation and azimuthal angles for the Matplotlib plot view. Default is
(22, 70).figsize (tuple, optional) – Figure size for the Matplotlib plot. Default is
(7, 4.5).text_wrap (int, optional) – Maximum width of the title text before wrapping. Useful for managing long titles. Default is
50.horizontal (float, optional) – Horizontal camera position for the Plotly plot. Default is
-1.25.depth (float, optional) – Depth camera position for the Plotly plot. Default is
1.25.vertical (float, optional) – Vertical camera position for the Plotly plot. Default is
1.25.cbar_x (float, optional) – Position of the color bar along the x-axis in the Plotly plot. Default is
1.05.cbar_thickness (int, optional) – Thickness of the color bar in the Plotly plot. Default is
25.title_x (float, optional) – Horizontal position of the title in the Plotly plot. Default is
0.5.title_y (float, optional) – Vertical position of the title in the Plotly plot. Default is
0.95.top_margin (int, optional) – Top margin for the Plotly plot layout. Default is
100.image_path_png (str, optional) – Directory path to save the PNG file of the Matplotlib plot. Default is None.
image_path_svg (str, optional) – Directory path to save the SVG file of the Matplotlib plot. Default is None.
show_cbar (bool, optional) – Whether to display the color bar in the Matplotlib plot. Default is
True.grid_resolution (int, optional) – The resolution of the grid for computing partial dependence. Default is
20.left_margin (int, optional) – Left margin for the Plotly plot layout. Default is
20.right_margin (int, optional) – Right margin for the Plotly plot layout. Default is
65.label_fontsize (int, optional) – Font size for axis labels in the Matplotlib plot. Default is
8.tick_fontsize (int, optional) – Font size for tick labels in the Matplotlib plot. Default is
6.enable_zoom (bool, optional) – Whether to enable zooming in the Plotly plot. Default is
True.show_modebar (bool, optional) – Whether to display the mode bar in the Plotly plot. Default is
True.
- Raises:
If
plot_typeis not one of"static","interactive", or"both".If
plot_typeis"interactive"or"both"andhtml_file_pathorhtml_file_nameare not provided.
- Returns:
NoneThis function generates 3D partial dependence plots and displays or saves them. It does not return any values.
Note
This function handles warnings related to scikit-learn’s
partial_dependencefunction, specifically aFutureWarningrelated to non-tuple sequences for multidimensional indexing. This warning is suppressed as it stems from the internal workings of scikit-learn in Python versions like 3.7.4.To maintain compatibility with different versions of scikit-learn, the function attempts to use
"values"for grid extraction in newer versions and falls back to"grid_values"for older versions.
3D Plots - CA Housing Example
Consider a scenario where you have a machine learning model predicting median
house values in California.[4]_ Suppose you want to understand how non-location
features like the average number of occupants per household (AveOccup) and the
age of the house (HouseAge) jointly influence house values. A 3D partial
dependence plot allows you to visualize this relationship in a more comprehensive
manner, providing a detailed view of how these two features interact to affect
the predicted house value.
For instance, the 3D partial dependence plot can help you explore how different combinations of house age and occupancy levels influence house values. By visualizing the interaction between AveOccup and HouseAge in a 3D space, you can uncover complex, non-linear relationships that might not be immediately apparent in 2D plots.
This type of plot is particularly useful when you need to understand the joint effect of two features on the target variable, as it provides a more intuitive and detailed view of how changes in both features impact predictions simultaneously.
Here’s how you can generate and visualize these 3D partial dependence plots using the California housing dataset:
Static Plot
Fetch The CA Housing Dataset and Prepare The DataFrame
from sklearn.ensemble import GradientBoostingRegressor
from sklearn.datasets import fetch_california_housing
from sklearn.model_selection import train_test_split
import pandas as pd
# Load the dataset
data = fetch_california_housing()
df = pd.DataFrame(data.data, columns=data.feature_names)
Split The Data Into Training and Testing Sets
X_train, X_test, y_train, y_test = train_test_split(
df, data.target, test_size=0.2, random_state=42
)
Train a GradientBoostingRegressor Model
model = GradientBoostingRegressor(
n_estimators=100,
max_depth=4,
learning_rate=0.1,
loss="huber",
random_state=1,
)
model.fit(X_train, y_train)
Create Static 3D Partial Dependence Plot
from eda_toolkit import plot_3d_pdp
plot_3d_pdp(
model=model,
dataframe=X_test,
feature_names_list=["HouseAge", "AveOccup"],
x_label="House Age",
y_label="Average Occupancy",
z_label="Partial Dependence",
title="3D Partial Dependence Plot of House Age vs. Average Occupancy",
image_filename="3d_pdp",
plot_type="static",
figsize=[8, 5],
text_wrap=40,
wireframe_color="black",
image_path_png=image_path_png,
grid_resolution=30,
)
Interactive Plot
from eda_toolkit import plot_3d_pdp
plot_3d_pdp(
model=model,
dataframe=X_test,
feature_names_list=["HouseAge", "AveOccup"],
x_label="House Age",
y_label="Average Occupancy",
z_label="Partial Dependence",
title="3D Partial Dependence Plot of House Age vs. Average Occupancy",
html_file_path=image_path_png,
image_filename="3d_pdp",
html_file_name="3d_pdp.html",
plot_type="interactive",
text_wrap=80,
zoom_out_factor=1.2,
image_path_png=image_path_png,
image_path_svg=image_path_svg,
grid_resolution=30,
label_fontsize=8,
tick_fontsize=6,
title_x=0.38,
top_margin=10,
right_margin=50,
left_margin=50,
cbar_x=0.9,
cbar_thickness=25,
show_modebar=False,
enable_zoom=True,
)
Warning
Scrolling Notice:
While interacting with the interactive Plotly plot below, scrolling down the page using the mouse wheel may be blocked when the mouse pointer is hovering over the plot. To continue scrolling, either move the mouse pointer outside the plot area or use the keyboard arrow keys to navigate down the page.
This interactive plot was generated using Plotly, which allows for rich, interactive visualizations directly in the browser. The plot above is an example of an interactive 3D Partial Dependence Plot. Here’s how it differs from generating a static plot using Matplotlib.
Key Differences
Plot Type:
The
plot_typeis set to"interactive"for the Plotly plot and"static"for the Matplotlib plot.
Interactive-Specific Parameters:
HTML File Path and Name: The
html_file_pathandhtml_file_nameparameters are required to save the interactive Plotly plot as an HTML file. These parameters are not needed for static plots.Zoom and Positioning: The interactive plot includes parameters like
zoom_out_factor,title_x,cbar_x, andcbar_thicknessto control the zoom level, title position, and color bar position in the Plotly plot. These parameters do not affect the static plot.Mode Bar and Zoom: The
show_modebarandenable_zoomparameters are specific to the interactive Plotly plot, allowing you to toggle the visibility of the mode bar and enable or disable zoom functionality.
Static-Specific Parameters:
Figure Size and Wireframe Color: The static plot uses parameters like
figsizeto control the size of the Matplotlib plot andwireframe_colorto define the color of the wireframe in the plot. These parameters are not applicable to the interactive Plotly plot.
By adjusting these parameters, you can customize the behavior and appearance of your 3D Partial Dependence Plots according to your needs, whether for static or interactive visualization.